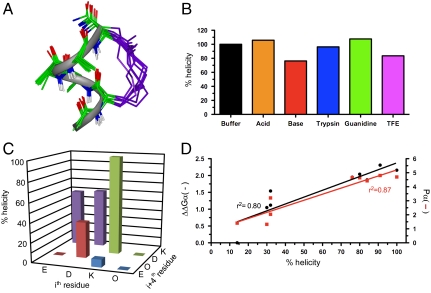
Fig. 1.
A stable α-helical turn tolerates amino acids from protein helices. (A) Compound 1, Ac-(1,5-cyclo)-[KAAAD]-NH2, is α-helical. Seventeen lowest energy NMR-derived structures for 1 in 90% H2O∶10% D2O at 298 K (backbone heavy atom pairwise rmsd 0.32 Å) showing α-helix (green), linker (purple), and three hydrogen bonds (dashes) vs. idealized α-helix (gray ribbon). (B) Compound 1 was stable (by circular dichroism changes at 222 nm after 30 min, 298 K) in aq. phosphate buffer (black), 1 M TFA (orange), 0.1 M KOH (red), 200 nM trypsin in 10 mM PBS at pH 7.4 (blue), 1∶1 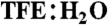 (purple), and 8 M guanidine (green). (C) Helicity depended on amino acid side chains at i and i + 4 positions that are covalently linked in cyclic peptides (2–9) Ac-(cyclo-1,5)-[(ith residue)-ARA-(i + 4 residue)]-NH2. The most α-helical had i = Lys, i + 4 = Asp. (D) % helicity in Ac-(2,6-cyclo)-R1[K2X3X4X5D6]-NH2 (10–18, Fig. S6) correlated both with the free energy (ΔΔGα, black) of each amino acid in stabilizing a protein α-helix (14) and with the probability (Pα, red) of an amino acid being in a protein α-helix (15). ΔΔGα and Pα values were summed for X3, X4, and X5 amino acids in each compound using literature data for each amino acid (14, 15). Helicity (%), ΔΔGα (kcal/mol), Pα (relative units) values are respectively 91, -2.31, 4.80 (10, A3A4A5); 100, -2.16, 4.70 (11, A3L4A5); 80, -2.04, 4.64 (12, A3M4A5); 84, -1.87, 4.42 (13, A3Q4A5); 76, -1.95, 4.65 (14, A3F4A5); 32, -1.54, 3.20 (15, A3G4A5); 32, -1.12, 2.04 (16, G3S4A5); 30, -1.05, 1.32 (17, S3S4S5); 14, 0.0, 1.41 (18, G3G4G5). Data for C and D were collected in 10 mM phosphate buffer (pH 7.4) at 25 °C (11).
(purple), and 8 M guanidine (green). (C) Helicity depended on amino acid side chains at i and i + 4 positions that are covalently linked in cyclic peptides (2–9) Ac-(cyclo-1,5)-[(ith residue)-ARA-(i + 4 residue)]-NH2. The most α-helical had i = Lys, i + 4 = Asp. (D) % helicity in Ac-(2,6-cyclo)-R1[K2X3X4X5D6]-NH2 (10–18, Fig. S6) correlated both with the free energy (ΔΔGα, black) of each amino acid in stabilizing a protein α-helix (14) and with the probability (Pα, red) of an amino acid being in a protein α-helix (15). ΔΔGα and Pα values were summed for X3, X4, and X5 amino acids in each compound using literature data for each amino acid (14, 15). Helicity (%), ΔΔGα (kcal/mol), Pα (relative units) values are respectively 91, -2.31, 4.80 (10, A3A4A5); 100, -2.16, 4.70 (11, A3L4A5); 80, -2.04, 4.64 (12, A3M4A5); 84, -1.87, 4.42 (13, A3Q4A5); 76, -1.95, 4.65 (14, A3F4A5); 32, -1.54, 3.20 (15, A3G4A5); 32, -1.12, 2.04 (16, G3S4A5); 30, -1.05, 1.32 (17, S3S4S5); 14, 0.0, 1.41 (18, G3G4G5). Data for C and D were collected in 10 mM phosphate buffer (pH 7.4) at 25 °C (11).