Abstract
Purpose
To review the existing endpoints of tumour growth delay assays in experimental radiobiology with an emphasis on their efficient estimation for statistically significant identification of the treatment effect. To mathematically define doubling time (DT), tumour-growth delay (TGD) and cancer-cell surviving fraction (SF) in vivo using exponential growth and regrowth models with tumour volume measurements obtained from animal experiments.
Materials and methods
A statistical model-based approach is used to define and efficiently estimate the three endpoints of tumour therapy in experimental cancer research.
Results
The log scale is advocated for plotting the tumour volume data and the respective analysis. Therefore, the geometric mean should be used to display the mean tumour volume data, and the group comparison should be a t-test for the log volume to comply with the Gaussian-distribution assumption. The relationship between cancer-cell SF, TGD and rate of growth is rigorously established. The widespread formula for cell kill is corrected; it has been rigorously shown that TGD is the difference between DTs. The software for the tumour growth delay analysis based on the mixed modeling approach with a complete set of instructions and example can be found on the author’s webpage.
Conclusions
The existing practice for TGD data analysis from animal experiments suffers from imprecision and large standard errors that yield low power and statistically insignificant treatment effect. This practice should be replaced with a model-based statistical analysis on the log scale.
Keywords: cell kill, doubling time, tumour-growth delay, surviving fraction
Introduction
There is an opinion that the most important part of experimental cancer research is the treatment regimen itself: If the effect of therapy is obvious, there is no need for complicated data analysis. This statement is hard to dispute. The problem is that often the effect is not so obvious. Moreover, it is usually obscured by notorious variation of animal response. Of course, one can detect a statistically significant treatment effect having perhaps hundreds animals per group. But our resources are limited and we typically have not more than 10 animals per group. That is why proper data analysis and clear definition of the endpoints of the treatment become as important as the treatment itself to prove its statistical significance.
There is much literature on the mathematical aspects of tumour growth using the theories of ordinary and partial differential equations (Banks 1994, Jones and Sleeman 2003). However these models contain so many unknown parameters that they become practically unidentifiable when fitted to typically short tumour-volume data. The tumour growth and regrowth model should be parsimonious in order to be practically useful. In this paper, we choose the exponential model, which can be adequate for the short period of tumour observation often used in experiments. In this work, we concentrate on the endpoints of tumour radiotherapy expressed in terms of the exponential growth and regrowth rather than complicated nonlinear models. Perhaps a more accurate analysis would be based on Gompertz’s model for tumour growth (Norton et al. 1976, Laird 1983, Castorina et al. 2007), but we deliberately choose the exponential growth model for the sake of transparency of the presentation.
The endpoints of experimental tumour biology and tumour growth delay assays particularly, including doubling time (DT) and tumour-growth delay (TGD), were discussed in qualitative form by such early authors as Begg (1980, 1987) and Hall (2000). However, there are only a few papers that discuss the statistical issues of TGD experiments (Bassukas 1993, Heitjan et al. 1993, Hanfelt 1997, Rygaard and Spang-Thomsen 1997). Remarkably, only a handful of tumour biology papers dealing with the analysis of growth-delay data cite these papers. Until now, the estimation of the DT and TGD in the majority of cancer research papers is carried out as suggested 40 years ago (Thomlinson and Graddock 1967, Denekamp 1974). In many papers, the determination of when the tumour reaches a specific volume is simply omitted. In several recent papers, the DT is computed based on measurements at two time points, one at time zero and at another arbitrary point (Ozono et al. 2004, Tomimaru et al. 2008). Some authors (Alley et al. 2004) advocate computation of the DT based on the group median growth, but then the standard errors required for the group comparison are not available. We question the validity of the popular t-test with the mean volume data because it violates the normal-distribution assumption.
The goal of the present paper is to advocate a model-based approach with clearly stated assumptions and rigorously defined endpoints using tumour-volume measurements in animal experiments. Usually, in those experiments several treatment groups are compared to find the most effective radiation or drug combination or to prove the efficacy of a new treatment regimen (Kallman 1987). We deliberately use a simple exponentialgrowth model, which is linear on the log scale. The three endpoints of tumour treatment are rigorously defined with discussion of how they can be efficiently estimated using modern statistical methodology. Practically, this methodology requires fewer animals per group to get statistically significant results compared with the traditional empirical approach.
The organisation of the paper is as follows: In the next section we review the major properties of the exponential growth and advocate the use of the log transformation of TGD data. Then three endpoints of the experimental cancer research, DT, TGD and surviving fraction (SF) are rigorously expressed using the exponential model-based approach. Finally, we illustrate the computation and interpretation of these end-points by an example of photodynamic radiotherapy.
Exponential tumour growth
In this section, we review some basic facts about exponential growth, emphasising the advantages of plotting the tumour volume data on the log scale. One may say that exponential growth can be observed only in untreated tumours, which usually constitute the control group, but we argue that the assumption of exponential growth is valid for treated tumours as well, after a certain period of time when such regrowth is evident.
The simplest model for tumour growth is exponential growth. This means that cells divide with a constant rate; the time between birth and division is constant. If V(t) denotes the tumour volume at time t, exponential growth implies
| (1) |
where V0 is the tumour volume at time t = 0 and β is the (relative) rate of tumour growth. We deliberately use a Greek letter, β, because the true rate of growth is typically unknown and estimated from the data; an estimate of β is denoted b, distinguishing between the true unknown value and its estimate. In particular, we interpret 100β% as the daily percent increase in tumour volume.
It is advantageous to plot the tumour growth data on the log scale because it turns exponential growth into linear growth:
where β is the slope and α = ln V0 is the intercept, the point where the straight line intersects the y axis. An important consequence for the data analysis is that an exponentially growing tumour volume plotted on the log scale looks like a straight line, which is easy to pick out by eye.
Usually, small tumours grow exponentially (after tumour vasculature is built), however the growth slows down when tumours get larger. Long ago, it was established that the Gompertz curve is an adequate model to capture the nonlinear dynamics of large tumours (Norton et al. 1976, Steel 1977, Laird 1983). Three special time points and corresponding tumour growth phases can be identified on the Gompertz curve based on the mathematical analysis of the rate of growth. The point of the fastest growth is where the first derivative is maximum (the second derivative vanishes). Using elementary calculus one can show that this time point occurs when tumour reaches 37% of its maximum volume. The points where tumour growth changes from slow to fast and back from fast to slow growth is where the third derivative is zero; as was shown in Demidenko (2004) these time points happen when tumour reaches 7.3% and 68% of its maximum volume respectively. Thus Gompertz growth has three phases: (a) The tumour development phase while vasculature is building – this phase takes the period from inoculation until the tumour reaches 7.3% of its maximum size; (b) the period of aggressive growth from 7.3–68% with the maximum rate of growth at 37% of its maximum volume; and finally (c) the period after the tumour reaches 68% when the growth is slowed by limited oxygen and nutrient supply.
In the majority of cases, cancer researchers deal with Phase 2: The tumour is not palpable in Phase I, and when the tumour becomes too large, it is either too late for a cure or the animal is sacrificed. Thus, the assumption of exponential growth may be valid in reality even for treated tumours after regrowth. We shall illustrate this assertion later.
Plotting tumour volume data
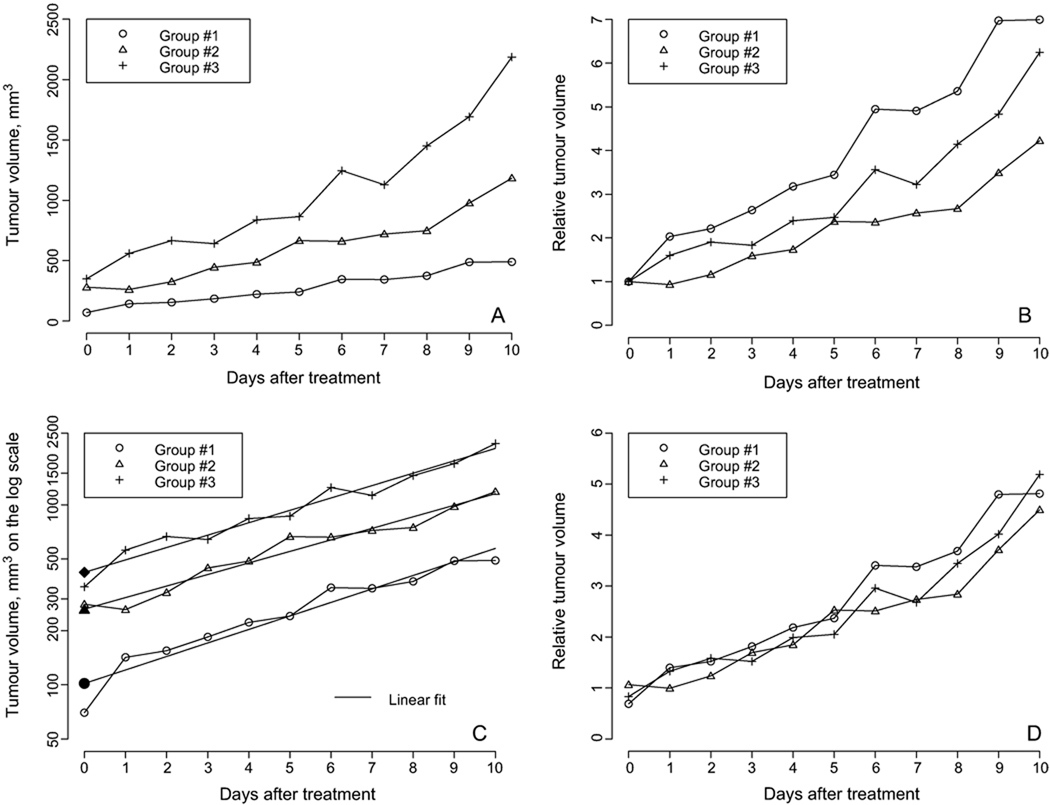
A visual representation of tumour-volume data is an important part of the analysis for treatment-group comparison. Three types of scales can be used: the original (linear) scale where the tumour volume is measured in cubic millimeters, the relative scale as the ratio of the tumour volume to the value at the starting time (t = 0), and the log scale. The three scales depicting somewhat typical tumour volume data in vivo with three treatment groups are illustrated in Figure 1 (radiation-induced fibrosarcoma tumours (RIF-1), a fast-growing subcutaneous tumour unpublished data obtained as a part of research reported in Hou et al. 2007).
Figure 1.
(A) Tumour volume growth in three groups on the original scale. It appears that the treatment effect is different in the different groups, the first group being the best. (B) The same tumour volume data but plotted on the relative scale. It seems that the first group is the worst in terms of the treatment effect. (C) Tumour volume data plotted on the log scale. The three groups are equivalent in terms of the rate of growth. The corresponding solid symbol depicts the estimate of the initial tumour volume from the linear fit. (D) The relative tumour volume for three treatment groups with the initial tumour volume estimated from the linear fit. In fact, the treatment effect in all three groups is the same.
Looking at the data on the original scale (Figure 1A), the reader may conclude that the three groups are different and the first group has the best treatment effect because the rate of tumour growth is smaller. However, one may notice that the initial tumour volume is different in the groups, so the relative tumour growth might seem beneficial (see Figure 1B). As follows from this graph, the first group has the worst treatment effect. A careful reader may notice that the reason why the first group data moved to the top is that the initial tumour volume is relatively low, so dividing the rest of the data by a smaller number moves the curve to the top. The log scale depicted in Figure 1C clarifies the group comparison: All three curves are close to straight parallel lines. This means that (a) the tumour volume grows exponentially in all three groups and (b) the rate of growth is almost the same in all cases. The final answer is that the treatment effect is the same in all three groups, at least visually.
The volume data of the exponentially growing tumours appear to be straight lines. If the lines for two treatment groups are parallel, we conclude that they have the same rate of growth.
Even when tumour growth has the same rate of growth, tumour growth curves plotted on the original scale may look different, so one may conclude that the treatment effect is different. Although the idea of eliminating the irrelevant differences in the initial volume is fruitful, the division operation may be confusing: if the initial volume is lower than expected, the entire curve moves up; if the initial volume is greater than expected, the entire curve moves down. Bassukas and Hofmockel (2001) also discouraged the use of relative tumour volume, advocating instead for Gompertz growth. We are not against using the relative tumour volume, we are against estimating the initial tumour volume as the mean volume at time zero. If data on the log scale look like a straight line, there is a more efficient way to estimate the initial tumour volume via the intercept from the linear model. Indeed, fitting the log tumour volume data to a linear function, α + βt, we obtain an estimate of the log initial tumour volume as a, the least squares estimate of the intercept α. In Figure 1C, the estimate of the log initial volume from the linear fit is depicted by a solid symbol. Now the relative tumour volume can be plotted either on the log scale as log Vt – a or on the relative scale as Vt/exp(a). The latter data are depicted in Figure 1D; it is clear that the three groups are equivalent in terms of tumour growth.
A hymn to the logarithm
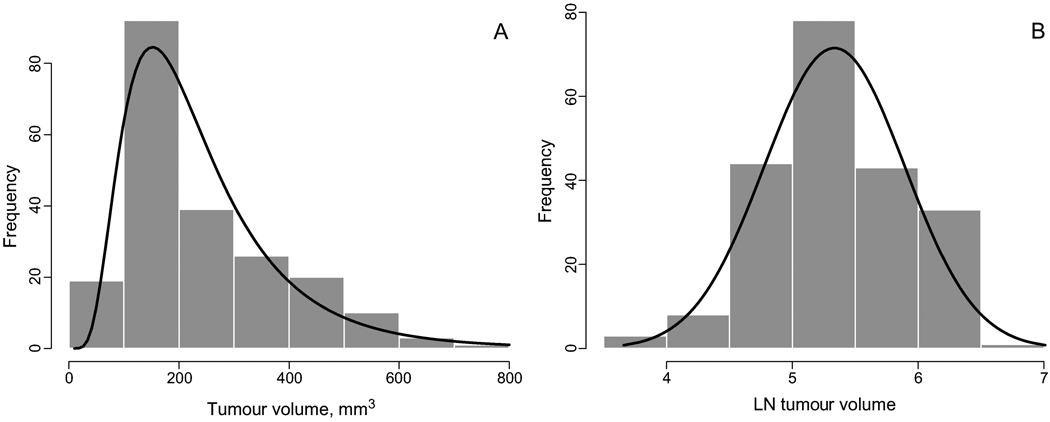
There is another good reason not only for plotting the data on the log scale, but for processing the data on the log scale. Most tumour volume data are skewed to the right: the number of animals with a tumour larger than the mean is greater than the number of animals with tumour volume less than the mean at every time point. In other words, the original tumour volume data are asymmetric and therefore do not comply with a Gaussian (or normal) distribution assumption. From statistics, we know that the arithmetic mean and standard deviation are efficient (have minimum variance) when the distribution is Gaussian; when the distribution is not Gaussian, there are more precise estimates of the theoretical mean and the standard deviation. A typical distribution of tumour volume data is shown in Figure 2; the actual data are taken from Zhou et al. (2006). The raw distribution (Figure 2A) is asymmetric and has a heavy right tail: The frequency of tumours with a larger volume is greater than that of tumours with a smaller volume.
Figure 2.
(A) The histogram and the estimated density of the tumour-volume data on the original scale. The distribution is asymmetric and skewed to the right (there are more larger tumours then smaller ones). (B) The histogram and the density of the same tumour volume data on the ln scale. The distribution is symmetric and is well approximated by the Gaussian density. The log transformation ‘makes’ the distribution of the tumour volume data Gaussian.
The log transformation eliminates skewness and makes the distribution of the tumour volume data much closer to the Gaussian distribution. This statement is illustrated in Figure 2B using the same data plotted in Figure 2A. After the log transformation, the data distribution becomes almost symmetric and is well described by the Gaussian density.
The fact that the original tumour volume data do not have a Gaussian distribution but the log data do has important implications in terms of computing the group mean and standard errors and deviations. As follows from statistical theory, the mean as the arithmetic average is an optimal estimator of the center of the distribution if the distribution is Gaussian. In particular, in the presence of the heavy right tail outliers may significantly affect the traditional mean with the bias toward large-volume outliers. The median is robust to outliers, but most statistical tests are developed for the mean under the assumption of a Gaussian distribution (there is no t-test for the median). Since the log data are well approximated by a Gaussian distribution, we need to compute the mean and standard deviation for the log data and then exponentiate them to get the optimal estimate on the original scale. As a result, the estimate of the group mean is the geometric mean, not the arithmetic mean. But it is a mathematical fact that the geometric mean is less than the arithmetic mean (equality holds if and only if all values are the same). This fact confirms our previous assertion that the traditional mean is positively biased toward larger volumes.
The doubling time (DT)
DT, defined as the time required to double the initial tumour volume, is a popular endpoint of experimental tumour radiobiology: the faster a tumour grows, the smaller DT. Using this definition, DT can be estimated either empirically by examining at what time the empirical tumour growth curve intersects the horizontal line 2V0, or from a model. Especially simple is computation of DT for an untreated, tumour growing exponentially according to formula (1). By the definition, DT is the solution to the equation V0eβt= 2V0 which implies:
| (2) |
Now we derive the formula for DT for treated tumours. We assume that a period of time after treatment, tregrowth, the tumour volume comes back to exponential growth with the same rate, β, as in the untreated group (we shall discuss this assumption in the Discussion section). Let this growth on the log scale be described as a linear function γ + βt where t ≥ tregrowth (due to treatment, we expect that γ < α). Then, by the definition, since V0 = eα the time t when the tumour volume doubles is found from 2eα = eγ+βt which implies the model-based formula,
| (3) |
Geometrically, DT is where the straight line of the tumour growth on the log scale, γ + βt, intersects the horizontal line α + ln 2. In practice, we replace the unknown rate of growth, β with its estimate b, and parameters α and γ with estimates a and g, respectively, from the fit by the linear function to the part of the data where the growth is exponential (see details in the example section below).
Sometimes, researchers work with the time when the treated tumour reaches triple the initial tumour volume. In that case, ln 2 should be replaced with ln 3.
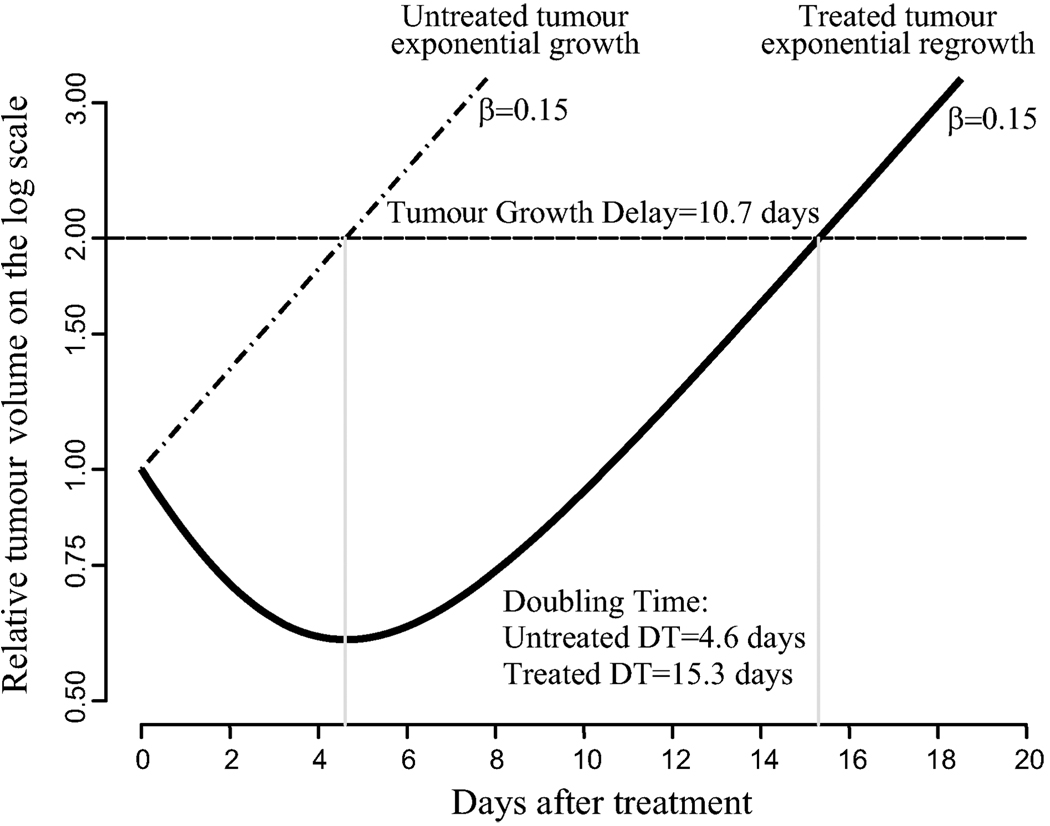
We illustrate the computation of DT in Figure 3, where the relative tumour volume (α = 0) is plotted versus time after treatment on the log scale. Without loss of generality, the treated and untreated tumour volumes can be set to 1. The untreated tumour grows exponentially with the rate β = 0.15 (15% volume increase per day). The DT is where the curve intersects the horizontal line V = 2 which gives DT = ln 2/0.15 = 4.6 days. The treated tumour shrinks after treatment, but then it regains the exponential growth with the same rate. As follows from this graph, the exponential growth returns starting at day tregrowth = 10. Fitting the linear model to this part of the data gives ν = −1.6 + 0.15t with DT as the solution to the equation − 1.6 + 0.15t = ln 2 which implies DT = (1.6 + ln 2)/0.15 = 15.3 days.
Figure 3.
A geometrical illustration of the computation of DT and TGD. It is assumed that after the treatment effect is over (in this case, approximately after day 10) the treated tumour regains its exponential growth with the same rate, β = 0.15, the lines are parallel on the log scale. TGD is the distance between lines on the time axis. DT is where the dashed horizontal line intersects the regrowth curve; approximately it can be computed from a linear equation.
In the model-based approach, DT is expressed through parameters of linear model on the log scale, and all tumour volume data are involved when these parameters are estimated from linear regression (see Example section for details). To the contrary, in the widespread empirical approach, DT is computed as the average over individual tumour growth curves, and only a couple of time points are involved close to double volume. There are several shortcomings to the empirical approach (non-model based) leading to a substantial bias and loss of statistical significance: (a) Some sort of interpolation is required because the horizontal line intersects the growth curve data between time points. (b) The approach is imprecise because of the impact of the initial tumour volume as in the calculation of the relative tumour volume. (c) The individual growth curve may be irregular (it can even intersect the horizontal line in two places), but a smoother mean curve does not allow the computation of the standard error of DT needed for a group comparison.
Tumour-growth delay (TGD)
TGD is a second popular endpoint of cancer radiotherapy. To simplify, we assume that untreated and treated tumours (after a period of time) grow exponentially with the same rate, see Figure 3 for an illustration. TGD is the distance on the x-axis between the two parallel straight lines. Following notation introduced in the previous section, we define TGD from solution of the equation:
which yields:
| (4) |
As with DT, one can demonstrate by statistical simulations that the empirical assessment of TGD is subject to large bias. TGD and DT are closely related. If a tumour regains its exponential growth after reaching the double volume, TGD is the difference in DT for treated and untreated tumours as is easy to see by taking the difference between expressions (3) and (2):
Also it is easy to see that TGD does not change if instead of DT one chooses the triple time. An extension of TGD to the Gompertz curve has been suggested in Bassukas (1993).
Cancer cell survival in vivo
The cancer cell survival assay is a common in vitro test of treatment efficacy in experimental cancer research (Sakamoto, 2004). This assay involves laborious procedures, but most importantly, its relation to in vivo cancer cell killing is unclear due to many principal differences of between the in vitro and in vivo environments. We assert that in vivo cancer cell survival can be estimated from the log tumour volume data. See Figure 4 for an illustration.
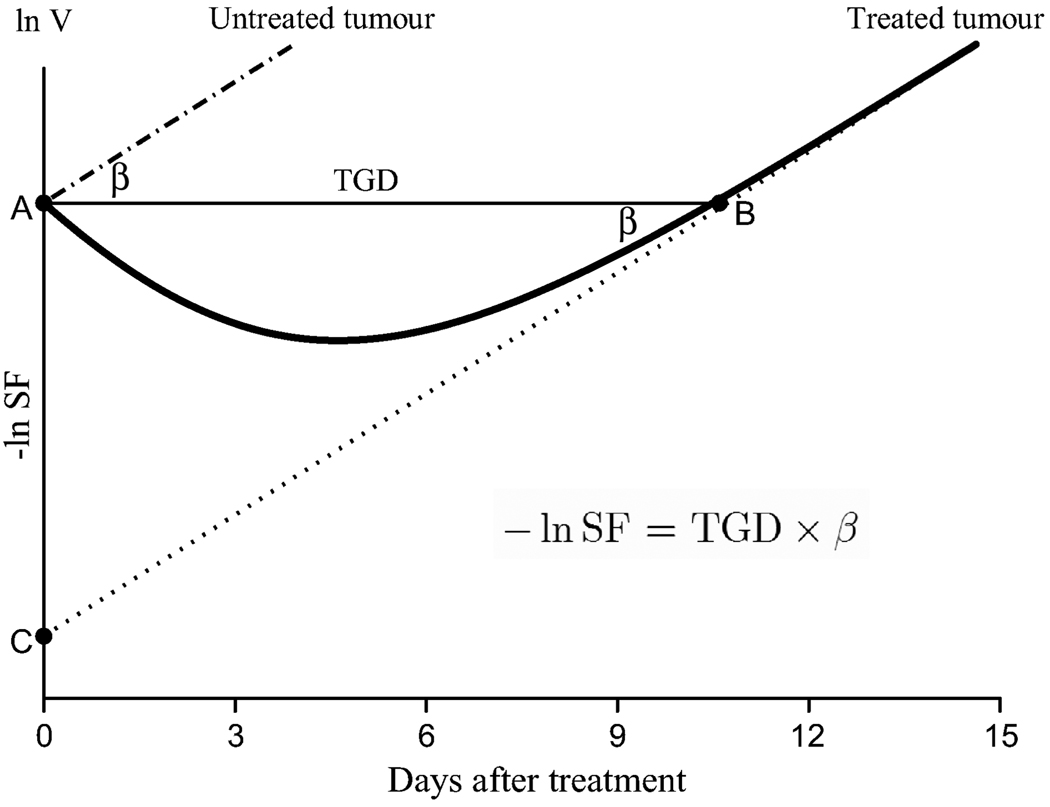
Figure 4.
A geometrical illustration of in vivo surviving fraction (SF) on the log scale. The log volume at time of the treatment (day = 0) is computed from the back projection of the regrowth line onto the y-axis. This population of cells represents surviving cells that grow exponentially with rate β, the dotted line. The relationship between the three endpoints of radiotherapy follows from the right triangle ABC.
As previously noted, it is assumed that untreated and treated tumours grow exponentially and, after a certain period of time, the treated tumour has the same rate of growth, β. In Figure 4, the regrowth with this rate starts at about 10 days after the treatment. Plotted on the log scale, both growth curves look like parallel straight lines. The x distance between the lines is TGD (side AB of the right triangle ABC). What population of cells at the time of the treatment (t = 0) would give rise to that regrowth (dotted line)? Clearly, we can find the answer by back projection onto the y axis, point C. If V0 is the initial tumour volume and S0 is the tumour volume that survives the treatment, the ordinate of point C is ln S0 the cell SF is S0/V0 and − ln SF = ln V0 − ln S0 In another interpretation, −ln SF, as the side AC of triangle ABC, is the distance between the untreated and treated tumour volume lines on the y axis. (The negative sign is used because SF < 1 and therefore ln SF < 0.) Combining these results, we finally obtain how the in vivo SF can be expressed via TGD and the rate of growth:
| (5) |
We can express SF and kill fraction (KF = 1 − SF) explicitly through TGD and β by exponentiating the above formula:
| (6) |
or equivalently:
| (7) |
where eγ is the population of cells/tumour volume that gave rise to exponential growth in the treated group with rate β after tregrowth.
It follows from these formulas that if TGD is close to zero, SF is close to 1 and KF is close to 0. Conversely, if the tumour growth delay is long. the cell KF is close to 1.
Several authors, including Papadopoulou et al. (2002) and Alley et al. (2004), use the following formula for cell kill suggested by Bissery et al. (1991):
| (8) |
where T and C are the median times in treated and control groups to reach a prespecified volume, respectively, and 0.301 = log102. In our opinion, this formula is incorrect, but nevertheless it has become popular among tumour biologists. Indeed, if (8) were correct, cell kill = 1 if there is no treatment (T = C). The correct formula for log KF using the previously derived formula (6), expressed in terms of T, C and DTuntr, is:
| (9) |
the proof is found in the Appendix (note that T and C are now model-based defined). As a simple check, when there is no treatment, the right-hand side of (9) is – infinity and the cell KF is zero (SF = 1).
Example: Photodynamic radiotherapy
In this section, we illustrate the statistical estimation of the three endpoints of experimental cancer research using model (1) in photodynamic therapy (PDT), an emerging cancer-treatment modality. We use the data from a study where the role of the photosensitiser was investigated using the mouse model with a flank-injected prostate tumour (Zhou et al. 2006). Tumour regrowth regained the original rate of growth by Day 4, so we analyse the data for t ≥ 4 in the treatment group. The data from the two groups are combined and estimated by one linear equation with a dummy variable j with j = 0 if the data are from the control/untreated group and j = 1 if the data are from the treated group after t = 3. Specifically, the following linear regression model is estimated.
where vijt is the log tumour volume of mouse i from group j at time tji and εijt error term with zero mean and constant variance. In this model, α is the log tumour volume at the beginning of the treatment (t = 0), β is the common rate of growth and δ = γ − α in the previous notation. Thus, we expect δ to be negative.
In a more sophisticated statistical model, it is assumed that the initial tumour volume is animal-specific which leads to
where bi represents the random effect term with zero mean and constant variance. This term explains the variation of the tumour volume at the time of treatment and in more general way reflects animal heterogeneity. The model with the random effect belongs to the family of linear mixed models (Demidenko 2004) and can be estimated using standard software, such as lme in the open-source statistical package R. The lme estimation results are presented in Table I.
Table I.
lme estimation results of ln tumour volume data in two groups in R.
| Parameter | Value | SE | Z-score | p-value |
|---|---|---|---|---|
| a (α) | 5.333 | 0.057 | 94 | <0.0001 |
| d (δ) | −2.188 | 0.096 | − 82 | <0.0001 |
| b (β) | 0.47 | 0.008 | 57 | <0.0001 |
| SDinit vol | 0.23 | |||
| SDerror | 0.19 |
The estimate of the overall slope coefficient (β) is b = 0.47, meaning that the tumour volume of mice from the control group and mice from the treated group after Day 4 grows exponentially at 47% a day. Intercept a = 5.333 estimates ln initial tumour volume at time 0 implying V0 = e5.333 = 207 mm3. For the treated group, intercept = 5.333 − 2.188 = 3.145 estimates the ln volume of cancer cells surviving the PDT treatment. In the bottom rows of Table I, we present the estimates of standard deviations of ln initial tumour volume and the error term. Remarkably, the deviation in the initial tumour volume is even larger than the deviation from the growth model. This confirms that for this application mixed model is more appropriate than traditional regression model which assumes that the variation of the initial tumour volume is zero. On the original scale, the standard deviations of the initial volume and error are 23% and 19%, respectively.
In Table II, we present the estimates of the three endpoints based on the results from Table I. The DT for the control group is computed by formula (2) with β = 0.47 and DT for the treated group is computed by formula (3) with a = 5.333 and g = 5.333 − 2.188 = 3.145 as an estimate of γ. Estimates of TGD and SF are computed using formulas (4) and (6), respectively, by replacing the true unknown values with their estimates. As follows from our calculations, the photodynamic radiation kills 89% of prostate cancer cells.
Table II.
Three endpoints in two mouse groups.
| Group | DT, days |
TGD, days |
ln SF | %SF | %KF | |
|---|---|---|---|---|---|---|
| Untreated/ Control |
|
0 | 0 | 100% | 0% | |
| SE | 0.026 | 0 | ||||
| PDT treated |
|
4.7 | −2.19 | 11.2% | 88.8% | |
| SE | 0.26 | 0.19 | 0.096 | 1.1% | 1.1% |
To test whether the rate of growth and regrowth is stable, we ran a mixed model with an individualspecific beta. The contribution of the variation of the initial tumour volume to the total variance was 85% while that of the variation of the rate, β, was 2%. Thus it is safe to assume that the rate of growth and regrowth is stable across individuals.
Discussion and summary points
We have rigorously defined and provided formulas for three endpoints of experimental cancer therapy using tumour-volume measurements in animal experiments – DT, TGD and cancer-cell SF – and established the relationships between them assuming exponential growth and regrowth. We emphasise that the empirical assessment of DT and TGD, widely used in practice among tumour biologists, is imprecise and biased, may lead to erroneous conclusions, and substantially reduces the power of establishing the statistical significance of the treatment effect. Of course, the model-based approach requires assumptions on the tumour growth and regrowth dynamics unlike the empirical approach. However in practice, there are simply not enough animal data to avoid those assumptions that substantially reduce statistical power for detection of the treatment effect.
We deliberately simplified our tumour regrowth model assuming that the rate of untreated tumour growth is the same as the rate of regrowth. We take this simplifying assumption so as to not bury principal concepts under technical issues and to make the presentation more accessible to the nonmathematician.
This paper makes five main points:
Tumour volume data should be plotted on the log scale: If the growth looks linear, the tumour growth is exponential; if the untreated and treated tumour growth lines look parallel, then their rates of growth are the same.
Computation of the group mean, standard deviation and t-test for group comparison should be done on the log scale because the log transformation eliminates skewness and makes the distribution Gaussian. Consequently, the geometric mean should be used instead of the arithmetic mean for the group mean tumour volume. The log transformation reduces the impact of large tumours/outliers and is better justified from a statistical point of view because the t-test assumes means, not medians.
TGD is the difference between the DT of treated and untreated tumours, assuming that by the time the tumour reaches the double volume, the rate of growth is the same as that of the untreated tumour (although TGD is usually computed as the difference between DTs, the assumption on equality of the growth rates is usually neither mentioned nor validated).
The formula for in vivo tumour cell kill, used in several papers, is incorrect. We provide a correct formula for in vivo cell SF expressed through tumour growth delay and rate of growth. Geometrically, TGD is the horizontal distance and −ln SF is the vertical distance between the straight lines on the log scale representing the treated and untreated tumour growth.
In order to efficiently estimate the endpoints of the TGD assay, one should apply a statistical model with data combined from different treatment groups using the dummy variable intercept term. Although such a model can be estimated using standard linear least squares, a more sophisticated mixed modeling technique is preferable. The random intercept model takes into account the heterogeneity of initial tumour volumes at the time of animal treatment.
One unwanted implication of our simplifying exponential-regrowth assumption is that the longitudinal data before regrowth are lost. More sophisticated, but nonlinear, regrowth models that describe the tumour volume dynamic on the entire time domain can be applied to eliminate this data loss (Demidenko 2004, 2006, Yu and Holmgren 2007).
Another limitation of the present approach is that the treatment is assumed to not affect the rate of regrowth. Although this is not a very serious assumption: all formulas can be accordingly modified even when the rates are not the same, except formula (4). Since the lines on the log scale are not parallel (usually regrowth is slower than growth), we still can compute and compare TGD, but then we need to specify the time at which the TGD is computed. The convenience of the parallel lines is that the TGD is time independent.
Our approach is applicable to fractionated and prolonged treatment regimens if the tumour regrowth with the initial rate of growth is regained after time tregrowth. Then the back projection of the regrowth straight line onto the y-axis estimates the log fraction of cells surviving with respect to the initial population of cells over the entire period of treatment. For example, in the case of fractionated radiation the SF computed by formula (6) and respectively the KF reflect the cumulative effect of treatment. However, this endpoint has a less clear interpretation than in the case of one treatment application because cell division and other longitudinal tumour changes over the course of treatment are involved. The formula for SF simply reflects the hypothetical proportion of cells that would give rise the exponential growth observed after tumour regains its initial rate of growth.
The existing practice in the statistical analysis of tumour growth delay data is fairly primitive and typically reduces to a t-test on the original scale. The empirical techniques to assess the DT and tumour growth delay were developed before computers were readily available and are still in use. These techniques are prone to bias and large standard errors. Model-based definitions of the endpoints of radiotherapy and their adequate statistical estimation using the mixed model is more powerful and requires fewer animals per group.
The complete set of instructions for the mixed model, the code and sample data can be found on the author’s website, www.dartmouth.edu/~eugened, accessible for non-statisticians. The output of that estimation procedure is a table similar to Table II.
Acknowledgements
The author would like to thank both reviewers for their helpful and constructive comments that improved the paper. This research was supported by grant R01 CA 130880 from NIH/NCI.
Appendix
Derivation of the cell-kill formula
We assume that the ln tumour volume in the control group grows as ν = α + βt for t ≥ 0 and in the treatment group as γ + βt after tregrowth where γ < α Let ν⋆ > α be the prespecified ln volume such that α + βtregrowth > ν⋆; we refer the reader to Figure 4 for an illustration. Then it is easy to see that the times to reach ν⋆ in the control and treatment groups are:
respectively. Plugging these formulas and formula (3) into (9), we obtain:
Now, using an elementary formula loga A = logb A/logb a, we represent log10 SF in the natural logarithm form as:
because log10 2/(ln 2 log10 e)= 1. This means that formulas (7) and (9) are equivalent. Since KF = 1 – SF, we express log10through the above formulas for C and T and formula (2) to obtain the final formula (9).
Footnotes
Declaration of interest: The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
References
- Alley MC, Hollingshead MG, Dykes DJ, Waud WR. Human tumor xenograft models in NCI drug development. In: Teicher BA, Andrews PA, editors. Anticancer drug development guide. 2nd ed. Totowa, NJ: Human Press; 2004. pp. 125–152. [Google Scholar]
- Banks RB. Growth and diffusion phenomena. Berlin: Springer-Verlag; 1994. [Google Scholar]
- Bassukas ID. A generalized tumor-growth delay assay for quantifying alterations of tumor regrowth patterns. Anticancer Research. 1993;13:1601–1606. [PubMed] [Google Scholar]
- Bassukas ID, Hofmockel G. Nobody needs “relative tumour sizes” to compare tumour growth curves. Anticancer Research. 2001;21:1181–1182. [PubMed] [Google Scholar]
- Begg AC. Analysis of growth delay data: Potential pitfalls. British Journal of Cancer. 1980;41:1639–1643. [PMC free article] [PubMed] [Google Scholar]
- Begg AC. Principles and practices of the tumor growth delay assay. In: Kallman RF, editor. Rodent tumor models in experimental cancer therapy. New York: McGraw-Hill; 1987. pp. 114–121. [Google Scholar]
- Bissery M-C, Guenard D, Gueritte-Voegeleina F, Lavelle F. Experimental antitumor activity of Taxotere (RP 56976, NSC 628503), a Taxol analogue. Cancer Research. 1991;51:4845–4852. [PubMed] [Google Scholar]
- Castorina P, Deisboeck TS, Gabriele P, Guiot C. Growth laws in cancer: Implication for radiotherapy. Radiation Research. 2007;168:349–356. doi: 10.1667/RR0787.1. [DOI] [PubMed] [Google Scholar]
- Demidenko E. Mixed models: Theory and applications. New York: Wiley; 2004. [Google Scholar]
- Demidenko E. The assessment of tumour response to treatment. Applied Statistics. 2006;55:365–377. [Google Scholar]
- Denekamp J. The response of a mouse sarcoma to single and divided doses of X-rays and fast neutrons. British Journal of Cancer. 1974;29:292–299. doi: 10.1038/bjc.1974.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall EJ. Radiobiology for the radiobiologist. 5th ed. Philadelphia: Lippincott; 2000. [Google Scholar]
- Hanfelt JJ. Statistical approaches to experimental design and data analysis of in vivo studies. Breast Cancer Research and Treatment. 1997;46:279–302. doi: 10.1023/a:1005946614343. [DOI] [PubMed] [Google Scholar]
- Heitjan DF, Manni A, Santen RJ. Statistical analysis of in vivo tumor growth experiments. Cancer Research. 1993;53:6042–6050. [PubMed] [Google Scholar]
- Hou H, Khan N, Grinberg O, Yu H, Grinberg S, Lu S, Demidenko E, Steffen RP, Swartz HM. The effects of Efaproxyn (efaproxiral) on subcutaneous RIF-1 tumor oxygenation and enhancement of radiotherapy-mediated inhibition of tumor growth in mice. Radiation Research. 2007;168:218–225. doi: 10.1667/RR0962.1. [DOI] [PubMed] [Google Scholar]
- Jones DS, Sleeman DS. Differential equations and mathematical biology. London: Chapman & Hall/CRC; 2003. [Google Scholar]
- Kallman RF. Rodent tumor models in experimental cancer therapy. New York: Pergamon Press; 1987. [Google Scholar]
- Laird AK. Dynamics of tumour growth. British Journal of Cancer. 1983;18:490–502. doi: 10.1038/bjc.1964.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton L, Simon R, Brereton HD, Bogden AE. Predicting the course of Gompertzian growth. Nature. 1976;264:542–545. doi: 10.1038/264542a0. [DOI] [PubMed] [Google Scholar]
- Ozono S, Miyao N, Igarashi T, Marumo K, Nakazawa H, Fukuda M, Tsushima T, Tokuda N, Kawamura J, Murai M. Tumor doubling time of renal cell carcinoma measured by CT: Collaboration of Japanese Society of Renal Cancer. Japanese Journal of Clinical Oncology. 2004;34:82–85. doi: 10.1093/jjco/hyh011. [DOI] [PubMed] [Google Scholar]
- Papadopoulou MV, Ji M, Ji X, Bloomer WD, Hollingshead MG. Therapeutic advantage from combining paclitaxel with the hypoxia-selective cytotoxin NLCQ-1 in murine tumor- or human xenograft-bearing mice. Cancer Chemotherapy and Pharmacology. 2002;50:501–508. doi: 10.1007/s00280-002-0521-8. [DOI] [PubMed] [Google Scholar]
- Rygaard K, Spang-Thomsen M. Quantitation and Gompertzian analysis of tumor growth. Breast Cancer Research and Treatment. 1997;46:303–312. doi: 10.1023/a:1005906900231. [DOI] [PubMed] [Google Scholar]
- Sakamoto K. Radiobiological basis for cancer therapy by total or half-body irradiation. Nonlinearity in Biology, Toxicology and Medicine. 2004;2:293–316. doi: 10.1080/15401420490900254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GG. Growth kinetics of tumours. Oxford: Clarendon Press; 1977. [Google Scholar]
- Thomlinson RH, Graddock EA. The gross response of an experimental tumour to single doses of X-rays. British Journal of Cancer. 1967;21:108–123. doi: 10.1038/bjc.1967.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimaru Y, Noura S, Ohue M, Okami J, Oda K, Higashiyama M, Yamada T, Miyashiro I, Ohigashi H, Yano M, Kodama K, Ishikawa O, Murata K, Yokouchi H, Sasaki Y, Kameyama M, Imaoka S. Metastatic tumor doubling time is an independent predictor of intrapulmonary recurrence after pulmonary resection of solitary pulmonary metastasis from colorectal cancer. Digestive Surgery. 2008;25:220–225. doi: 10.1159/000140693. [DOI] [PubMed] [Google Scholar]
- Yu RX, Holmgren RX. Endpoints for agents that slow tumor growth. Contemporary Clinical Trials. 2007;28:18–24. doi: 10.1016/j.cct.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Zhou XD, Pogue BW, Chen B, Demidenko E, Joshi R, Hoopes J, Hasan T. Pretreatment photosensitizer dosimetry reduces variation in tumor response. International Journal of Radiation Oncology Biology Physics. 2006;64:1211–1220. doi: 10.1016/j.ijrobp.2005.11.019. [DOI] [PubMed] [Google Scholar]