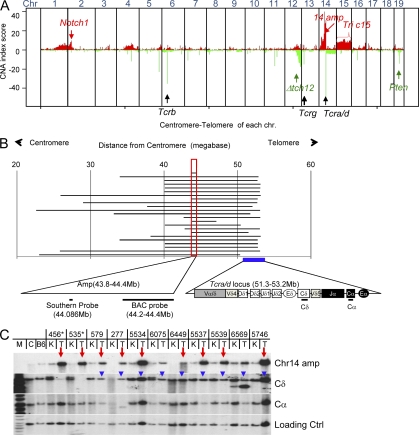
Figure 1.
Amplification of sequences centromeric to the Tcra/d locus in Atm−/− T cell lymphomas. (A) Combined CGH analysis of 18 ATM-deficient thymic lymphomas. Each tumor sample was hybridized and analyzed once with matched kidney control DNA. The y axis represents amplification/deletion index (sum of LN tumor/kidney ratio of all tumors analyzed). Amplification and deletion were scored separately and plotted together. The amplification score at each probe location is plotted in red and deletion score is plotted in green. At some loci, such as the telomeric end of chromosome 12, both deletions and amplifications were found in this collection of tumors, which is termed copy number polymorphism, indicating inconsistent copy number changes in this region. The x axis depicts all 19 mouse chromosomes (1 to 19 arranged from centromere to telomere). The red arrows and text highlight recurrent amplifications of Notch1 and chromosome 14 sequences upstream of the Tcra/d locus, and as well as recurrent trisomy 15 (Tric15). The black arrows and text indicate the position of Tcr gene loci. The green arrows and text highlight recurrent deletions in the telomeric portion of chromosome 12 (Δtch12) and the Pten locus. (B) The diagram shows the length of amplification (defined by LN2 tumor/kidney > 0.667) in each of the ATM-deficient thymic lymphomas. The x axis is the base pair position indicated by distance (Mb) from the chromosome 14 centromere. The common amplification region is marked with a red box and shown in the expanded form at the bottom. The position of the BAC probe used for FISH (RP23_359O5) and probe used for Southern blotting are indicated. The Tcra/d locus is indicated with a filled blue box and expanded at the bottom of the panel to show relative orientation. (C) Southern blot analyses of 11 tumor/kidney DNA pairs. About 10 µg of genomic DNA from kidney (K) and the corresponding tumors (T) was digested with EcoRI and probed with the probe described in B (Chr14 Amp) and with probes that recognize the Tcrd constant region (Cδ) or the Tcra (Cα). The red arrows indicate tumor samples with significant amplification. The blue arrowheads indicate the retention of Cδ in those tumors. The asterisks mark the two samples that show Cδ deletion as predicted by CGH analysis (Fig. 3 B). The loading control probe was derived from an intron of the Mdc1 gene (which lies on mouse chromosome 17). M, DNA Marker; C, control DNA from 129sv mouse strain; B6, control DNA from BL6 mouse strain.