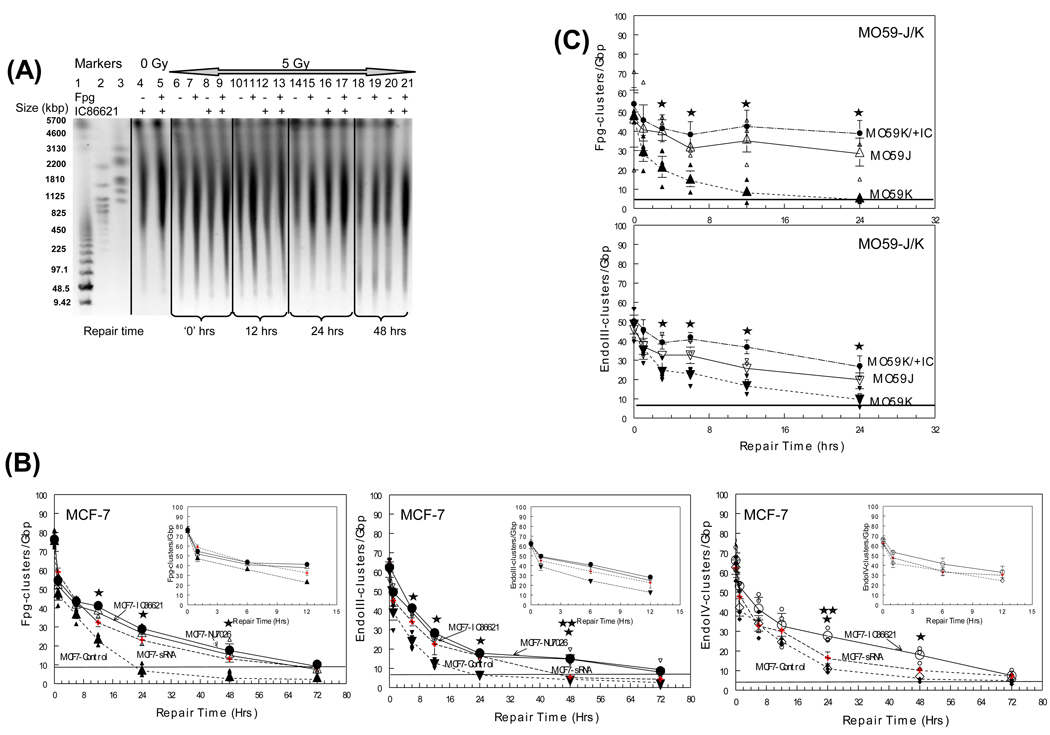
Fig. 2.
Processing of non-DSB oxidative DNA clusters in DNA-PKcs proficient and deficient cell lines (IC86621-, NU7026-, siRNA-treated MCF-7 and MO59-K, MO59-J and MO59-K) as a function of post-irradiation time. Different types of damage clusters were detected using PFGE analysis and E. coli repair enzymes Fpg and EndoIII as damage probes: (A) Representative image of a neutral gel containing DNA from MCF-7 cells unirradiated (0 Gy) and exposed to 5 Gy of γ rays, and harvested as a function of repair time post-exposure. Molecular size markers, lanes 1–3: S. pombe and low range PFG marker, lane 1; H. wingei, lane 3; S. cerevisae, lane 3. The molecular sizes of these DNAs are shown in kbp at the left of the gel image; 0 Gy, lanes 4–5; 5 Gy at increasing repair times (0–72 hrs), lanes 6–21. For each time point four samples are shown: enzyme or IC86621 untreated, (−); Fpg- or IC86621-treated, (+). (B–C) Cluster repair detected in MCF-7 and MO59-J/K cells. Fpg-detected oxypurine clusters, upright triangles. Endo III-detected oxypyrimidine clusters, inverted triangles. Endo IV detected abasic sites, circles. Values for DNA-PK siRNA cells (up to 24 hrs) are taken from Peddi et al. [29] to allow direct comparison while for 48 and 72 hrs are from this study (crosses). Values are averages from three (MCF-7) or two (MO59-J/K) independent experiments. Insets: Average values for MCF-7 with repair times up to 12 hr. Small symbols, individual data points from independent irradiation experiments; Large symbols, averages. Error bars, SEM; in some cases are smaller than the corresponding symbol. Closed symbols, control MCF-7 or MO59-K cells. Closed large circles, NU7026-MCF-7 or IC86621-MO59K (MO59K/+IC). Open symbols, IC86621-treated MCF-7 cells or M059-J. Statistically significant differences between IC86621-/NU7026-treated MCF-7 or MO59-K (MO59K/+IC) and controls (*) and between IC86621-/NU7026-MCF-7 and siRNA-MCF-7 (**) at p<0.05 are shown. Straight lines correspond to sham-irradiated (0 Gy) average values for each type of cluster.