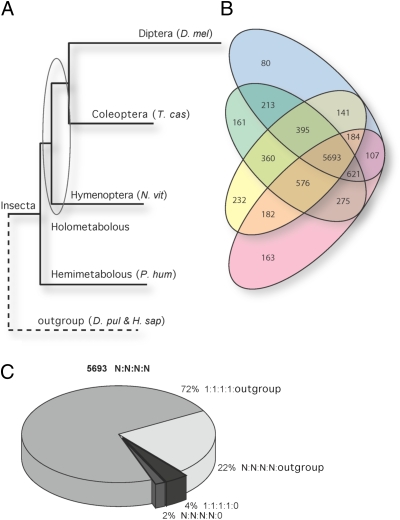
Fig. 1.
The Pediculus humanus humanus (P. hum) genome reveals a basal insect gene repertoire. The encoded P. hum proteome is compared with sequenced representatives of the orders Diptera, Coleoptera, and Hymenoptera and outgroup species beyond Insecta. D. mel, Drosophila melanogaster; T. cas, Tribolium castaneum; N. vit, Nasonia vitripennis; D. pul, Daphnia pulex; H. sap, Homo sapiens. (A) The Maximum-Likelihood phylogenetic tree was reconstructed using the superalignment of protein sequences of universal single-copy orthologs. The obtained tree confirms the basal position of Hemimetabola compared with Holometabola within Insecta. The branch lengths are proportional to the accumulated number of substitutions, suggesting an average rate of molecular evolution in lice that is comparable with that in Hymenoptera and Coleoptera. (B) The Venn diagram shows the numbers of orthologous groups of genes shared among the four insects (a lower estimate of the ancestral number of genes). It depicts the phylogenetic distribution of orthologs, highlighting the completeness of the gene repertoire encoded in the body louse genome. Pink, P. hum; yellow, N. vit; green, T. cas; blue, D. mel. (C) The pie chart partitions the largest fraction of core body louse proteins with orthologs in three holometabolous insect orders and the outgroup species beyond Insecta with respect to single- (1:1:1:1) and multiple- (N:N:N:N) copy orthologs. Of 5,693 groups of single- and multiple-copy orthologs common across Insecta, 94% are shared across Bilateria as single-copy (72%) or multiple-copy (22%) orthologs, and only 6% are insect-specific orthologous groups (4% as single copies and 2% as multiple copies).