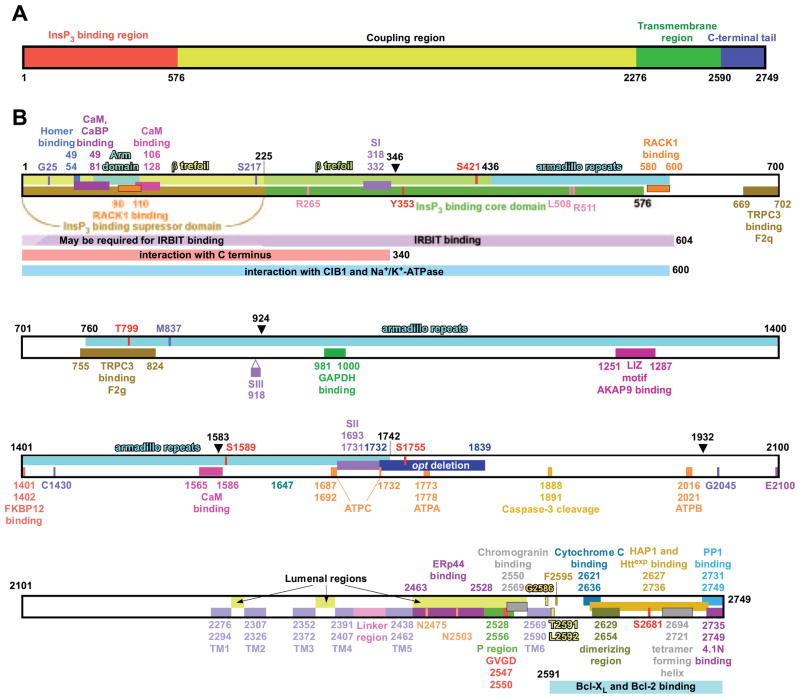
FIG. 2.
Structural determinants of the InsP3R. A: overall domain structure. The InsP3R molecule depicted as a linear amino acid sequence, with the NH2-terminal InsP3 binding region (red), coupling region (yellow), transmembrane region (green), and COOH tail (blue) depicted. B: linear amino acid sequence. Residues are numbered according to the rat type 1 SI+, SII+, SIII− sequence (protein accession no. 121838). Structural features (see section in this review where each element is described) shown are as follows: arm subdomain and β-trefoil in the InsP3-binding suppressor domain (sect. IIIB1); β-trefoil and armadillo repeats in InsP3 binding core domain (sect. IIIB1); armadillo repeats in the coupling domain (sect. IIIB3); alternative splicing regions SI, SII, and SIII (sect. IIB2) for type 1 InsP3R; opt deletion in type 1 InsP3R mutant (sect. IIIB3); ATP-binding sites ATPA, ATPB, and ATPC (sect. VIF4); transmembrane helices TM1–6 and pore-forming P region (sect. IIIB2A) with selectivity filter (sect. VF); linker region (sect. IIIB1); dimerizing region (sect. IIIC1); tetramer forming region (sect. IIIC1). Trypsin proteolysis sites (sect. IIIC3) are indicated by black arrowheads. The caspase 3 cleavage site (sect. IIIC3) is also shown. G25 (sect. IIIB1), S217 (sect. IIIB1), T799 (sect. IIIB3), M837 (sect. IIIB3), C1430 (sect. IIIB3), and G2045 (sect. IIIB3) are highly conserved residues the mutation of which can impact InsP3R channel functions. Mutation of E2100 modifies Ca2+ regulation of InsP3R channel (sect. VIK). R265, L508, and R511 are critically important for InsP3 binding (sect. IIIB1). G2586, T2591, L2592, and F2595 are residues that may be involved in forming the gate of the InsP3R channel (sect. IIIB2B). N2475 and N2503 are glycosylation sites (sect. IIIB). S1589 and S1755 are PKA/PKG phosphorylation sites (sect. VIL, 1 and 2); S2681 is an Akt phosphorylation site (sect. VIL4); Y353 is a Fyn tyrosine kinase phosphorylation site (sect. VIL5); and S421 and T799 are cdc2/CyB phosphorylation sites (sect. VIL6). Sequences involved in interaction of InsP3R channel with the following proteins are also depicted: homer (sect. VIN13); calmodulin (sect. VIN1); CaBP (sect. VIN2); RACK1 (sect. VIN3); IRBIT (sect. VIN4); CIB1 (sect. VIN2); Na+-K+-ATPase (sect. VIN14); COOH terminal of InsP3R in the tetrameric channel (sect. IIIB1); TRPC3 (sect. VIN14); GAPDH (sect. VIN8); AKAP9 [through leucine/isoleucine zipper (LIZ) motif] (sect. VIL1); FKBP12 (sect. VIN7); Erp44 (sect. VIN6); chromogranins (sect. VIN5); cytochrome c (sect. VIN10); HAP1 and Httexp (sect. VIN11); protein 4.1N (sect. VIN13); and PP1 (sect. VIL1).