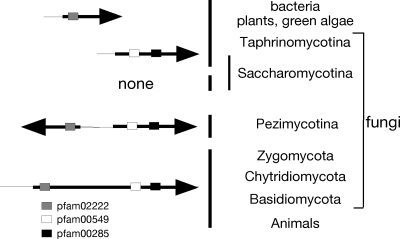
Fig. 2.
ACL gene arrangements in different phyla. See reference 14 for information on bacteria, algae, plants, and animals. BlastP searches of fungal genomes (http://www.broadinstitute.org/annotation/fungi/index.html) using the predicted A. nidulans ACL-encoding genes AN2435.3 and AN2436.3 showed predicted divergently transcribed genes in the genomes of all species of the Pezimycotina, e.g., Magnaporthe grisea (MGG_06720.5 and MGG_06719.5), Scelerotinia sclerotiorum (SS1G_02378.1 and SS1G_02379.1), and Botrytis cinerea (BC1G_05991.1 and BC1G_05989.1). In the case of Neurospora crassa (NCU06783 and NCU06785), a very short dubious predicted gene (NCU06784) occurs between the genes. Single genes are found in Zygomycota (Rhizopus oryzae [RO3G_14197.1]), Chytridiomycota (Batrachochytrium dendrobatidis [BDEG_06723]), and Basidiomycota (e.g., Ustilago maydis [UM01005.1], Cryptococcus neoformans [CNAG_04640.1], and Coprinus cinerea [CC1G_09654.1]). Analysis of fungal orthogroups (http://www.broadinstitute.org/regev/orthogroups/) showed no ACL homologues in the Saccharomycotina, with the exception of Y. lipolytica (YALI0D24431g and YALI0E34793g). Schizosaccharomyces pombe (Taphrinomycotina) contains two unlinked ACL genes (SPBC1703.07 and SPAC22A12.16). Conserved protein domains (http://www.ncbi.nlm.nih.gov/sites/entrez) are shown: pfam02222 (ATP grasp domain), pfam 00549 (CoA ligase), and pfam00285 (citrate synthase).