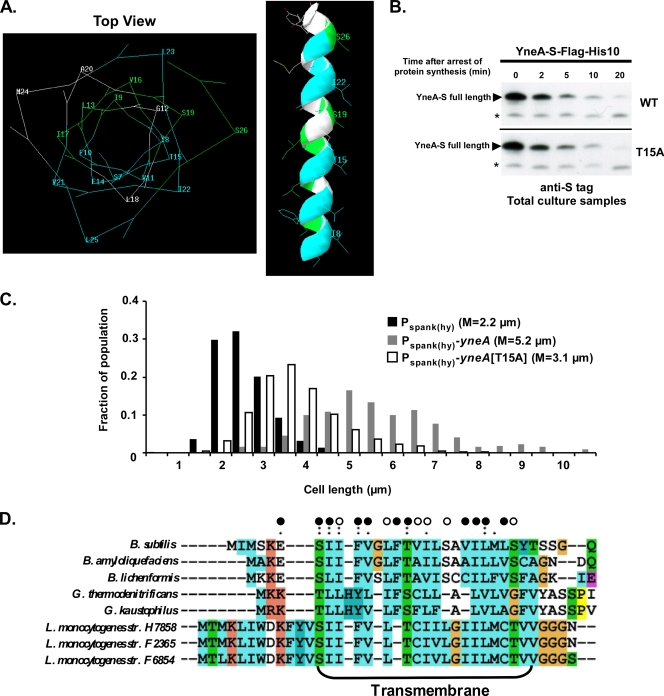
FIG. 7.
Mutations in the transmembrane region inactivate YneA. (A) Mutations that inactivate YneA cluster on one face of the predicted transmembrane region. The transmembrane region was modeled as an alpha-helix, using Swiss-PDP Viewer. Residues are colored as follows: blue indicates alanine substitutions that inactivate YneA, green indicates alanine substitutions that have little or no effect on YneA, and white indicates residues that were not mutated or tested. (B) The T15A mutation does not affect the stability of YneA. AM199 [thrC::Pspank(hy)-yneA-S-flag-his10] and AM261 [thrC::Pspank(hy)-yneA(T15A)-S-flag-his10] were grown and samples were prepared as described in the legend to Fig. 5D. Proteins were separated in a 13.8% SDS-polyacrylamide gel, and immunoblots were performed using diluted (1:5,000) anti-S primary antibody. Asterisks indicate nonspecific bands. (C) The T15A mutation reduces YneA function. AM69 [amyE::Pspank(hy)-yneA], AM70 [amyE::Pspank(hy)], and AM239 [amyE::Pspank(hy)-yneA(T15A)] were grown and induced (in the same experiment as that for Fig. 6B), and cells were stained and measured as described in the legend to Fig. 1C. This experiment is representative of two independent trials. (D) Many inactivating mutations are in conserved residues. Alignment of transmembrane domains in YneA orthologues was performed using ClustalX2.0. Open circles, alanine substitutions with little or no effect on YneA activity; closed circles, alanine substitutions that inactivate YneA; single and double dots indicate conserved amino acid groups.