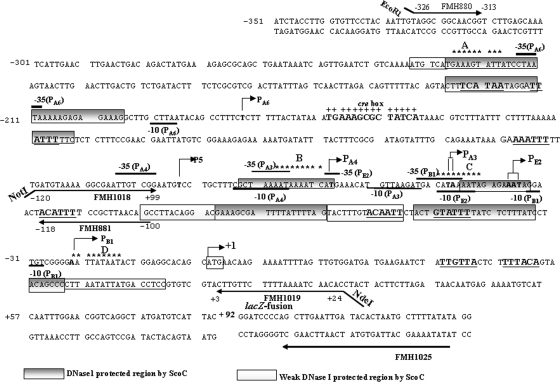
FIG. 1.
The phoPR promoter sequence and 5′ PhoP coding sequence showing six transcription start sites along with the putative ScoC binding sites. Transcription start sites for PB1, PE2, PA3, PA4, P5, and PA6 are indicated by boldface sequence and are identified by a bent arrow followed by the promoter number and a letter identifying the form of RNAP (where known) required for the transcription. The −10 region is marked below, and the −35 is marked above the sequence for each promoter based on published consensus sequences (16). The region of +1 to +92 is the 5′ PhoP coding sequence followed by the lacZ fusion. The translation start site, ATG, is boxed and identified by a bent arrow marked +1. Sequence numbering is relative to the A of ATG as +1. A single putative ScoC binding site in the 5′ region of the phoPR promoter, 5′-−227GAAAGTATT−219-3′ (site A), is located on the coding strand. Three conserved putative ScoC binding sites in the 3′ region of the phoPR promoter are 5′-78AATAAAATC−71-3′ (site B), 5′-−51CATAAAATA−43-3′ (site C), and 5′-−23AATTATAAT−15-3′ (site D) located on the coding strand. The ScoC binding consensus sequence is RATANTATY, where R is A or G, Y is C or T, and N is A, G, C, or T. The putative ScoC binding sites are indicated with nine stars above the sequences. The CcpA binding site (cre box) is shown in bold, marked above with plus signs. Primers used are shown as underlined or overlined sequences with an arrow in the direction of synthesis. FMH1025 is the primer used for primer extension analysis specific to the lacZ fusion. The ScoC DNase I-protected regions are shown as gray boxes on both strands of the phoPR promoter. The white boxes show the weaker ScoC protection. The consensus repeats for PhoP dimer binding, TT(A/C/T)A(C/T)A, are underlined, and the sequence is in boldface.