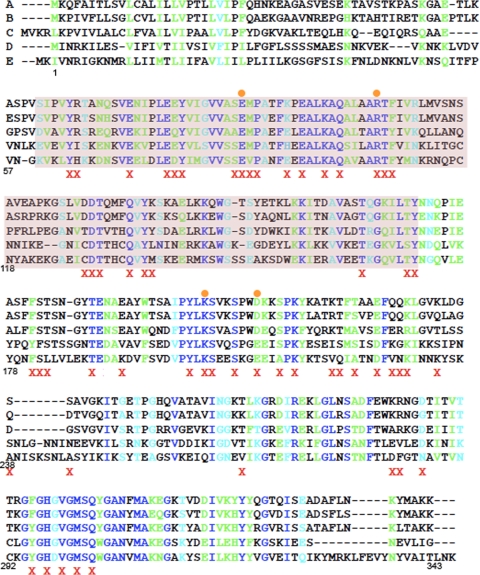
FIG. 2.
Amino acids selected for mutagenesis of SpoIID. Alignments of spoIID were performed by using CLUSTAL W (21). The species presented here are as follows: A, Bacillus subtilis; B, Bacillus amyloliquefaciens; C, Geobacillus thermodinitrificans; D, Clostridium perfringens; E, Clostridium saccharobutylicum. Amino acid numbers below the alignment refer to the B. subtilis sequence for easier identification of mutations. A red X indicates an amino acid changed to alanine and an orange circle indicates a mutant characterized in the present study. Sequence homology is represented by color, with completely conserved amino acids indicated by royal blue, highly conserved amino acids indicated by green and weakly conserved amino acids indicated by cyan. The putative peptidoglycan degradation domain is highlighted in purple. Recently, an alignment and compilation of all 667 SpoIID family members has become available on Pfam (14). This alignment was used to confirm our earlier alignments and verify that all residues showing significant conservation were mutated during our study. In this more complete alignment, only six amino acids are completely conserved, and just two of these, E88 and R106, resulted in an in vivo phenotype when changed to alanine.