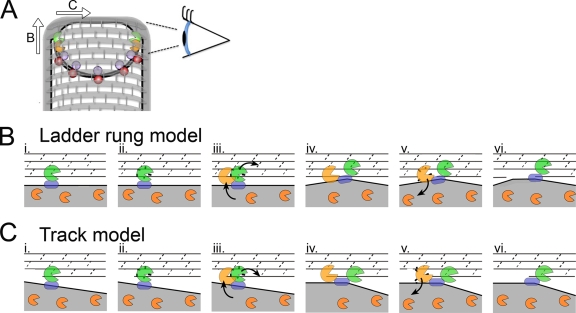
FIG. 7.
Model for SpoIIDMP function in vivo. (A) An engulfing B. subtilis cell with SpoIID (orange), SpoIIP (green), SpoIIQ (purple), and SpoIIIAH (red) indicated. Peptidoglycan is drawn with gray hoops representing glycan strands and short gray rods representing peptide cross-links. The eye orients the reader to the perspective used in panels B and C. As indicated by the arrows, the DMP protein complex can move in one of two directions relative to the long axis of the cell. If the proteins move from one glycan chain to the next (as shown in panel B and proposed by Morlot et al. [27]), each complex would move along the long axis of the cell. If the proteins travel along one glycan chain (as shown in panel C and proposed by Abanes-De Mello et al. [1]), then each complex would move across the short axis of the cell. (B and C) Top-down views of the leading edge of the engulfing mother cell membrane (gray fill) advancing over the forespore peptidoglycan, drawn with gray lines representing the glycan chains and dashed lines representing the peptide cross-links. (B) Ladder rung model, in which SpoIIP moves from one glycan chain to the next. (i) SpoIIP (green) is bound in the membrane by SpoIIM (blue) and to peptidoglycan. SpoIID (orange) is free in the membrane. (ii) SpoIIP cleaves peptide cross-links. (iii) SpoIID is recruited to the recently denuded glycan strands where it displaces SpoIIP to a new target on the next strand, laddering up the peptidoglycan. (iv) Both enzymes are in complex and bound to the peptidoglycan. (v) SpoIID cleaves the glycan strand, loses affinity for either SpoIIP or the peptidoglycan, and exits the complex. (vi) Same as panel i, but membranes have advanced. (C) Track model, similar to panel B, but depicting SpoIIDMP moving along the glycan chain, such that at step iii, SpoIID displaces SpoIIP to the next peptide cross-link on the same glycan strand.