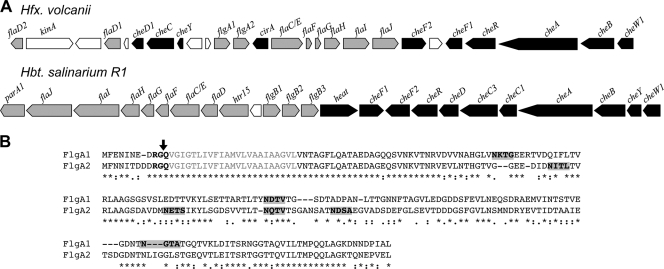
FIG. 1.
H. volcanii flgA1 and flgA2 genomic loci and amino acid sequences of FlgA1 and FlgA2. (A) H. volcanii motility and chemotaxis gene cluster compared to that of Halobacterium salinarum R1 as described by Schlesner et al. and the HaloLex Web server (25, 47, 48). The arrows represent the open reading frames (ORFs) encoding H. salinarum R1 and H. volcanii homologs of motility (gray) and chemotaxis (black) genes. The orientation of the arrows corresponds to the orientation of the ORFs relative to other genes within the putative operons. Each H. volcanii gene is annotated based on significant BLASTP alignment scores with experimentally verified homologous proteins. White arrows represent ORFs of unknown function. H. volcanii genes correspond to Hvo_1200 (flaD2) to Hvo_1225 (cheW1). (B) Amino acid alignment of the flagellins FlgA1 and FlgA2, as determined using ClustalW2 (32). Asterisks indicate identical residues, colons indicate conserved substitutions, and periods indicate semiconserved substitutions. The PibD-processing motif (bold) was predicted by FlaFind (52); the arrow shows the predicted PibD cleavage site. Signal peptide hydrophobic stretches (gray) were predicted by TMHMM (50). Putative glycosylation sites of mature FlgA1 and FlgA2 (bold and shaded gray) were predicted using the NetNGlyc, version 1.0, server (http://www.cbs.dtu.dk/services/NetNGlyc/).