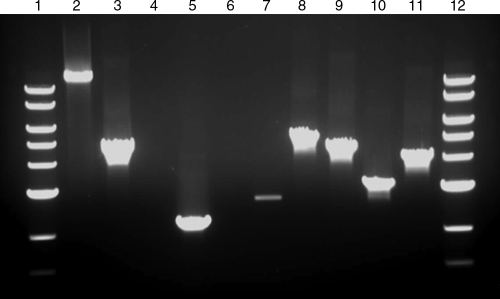
FIG. 4.
PCRs used to confirm erythromycin marker integration in the T. saccharolyticum genome. Lane 1, NEB 10-kb ladder; lane 2, wild type with T4P external primers X08727, X08728, predicted size 12,981 bp; lane 3, M1464 with T4P external primers, predicted size 4,669 bp; lane 4, wild type with cinA recA downstream external primer X08736 and erythromycin internal primer X00957, no predicted band; lane 5, M1465 with primers X08736 and X00957, predicted size 2,191 bp; lane 6, wild type with cinA recA upstream external primer X08735 and erythromycin internal primer X00958, no predicted band; lane 7, M1465 with primers X08735 and X00958, predicted size 2,692 bp (internal and external primers were used to verify M1465, as the erythromycin gene replaced a similarly sized fragment of the cinA recA locus); lane 8, wild-type with comEC external primers X08160 and X08161, predicted size 5,217 bp; lane 9, M1466 with comEC external primers, predicted size 4,519 bp; lane 10, wild type with comEA external primers X08154 and X08155, predicted size 3,165 bp; lane 9, M1467 with comEA external primers, predicted size 4,187 bp; lane 12, NEB 10-kb ladder.