Figure 2.
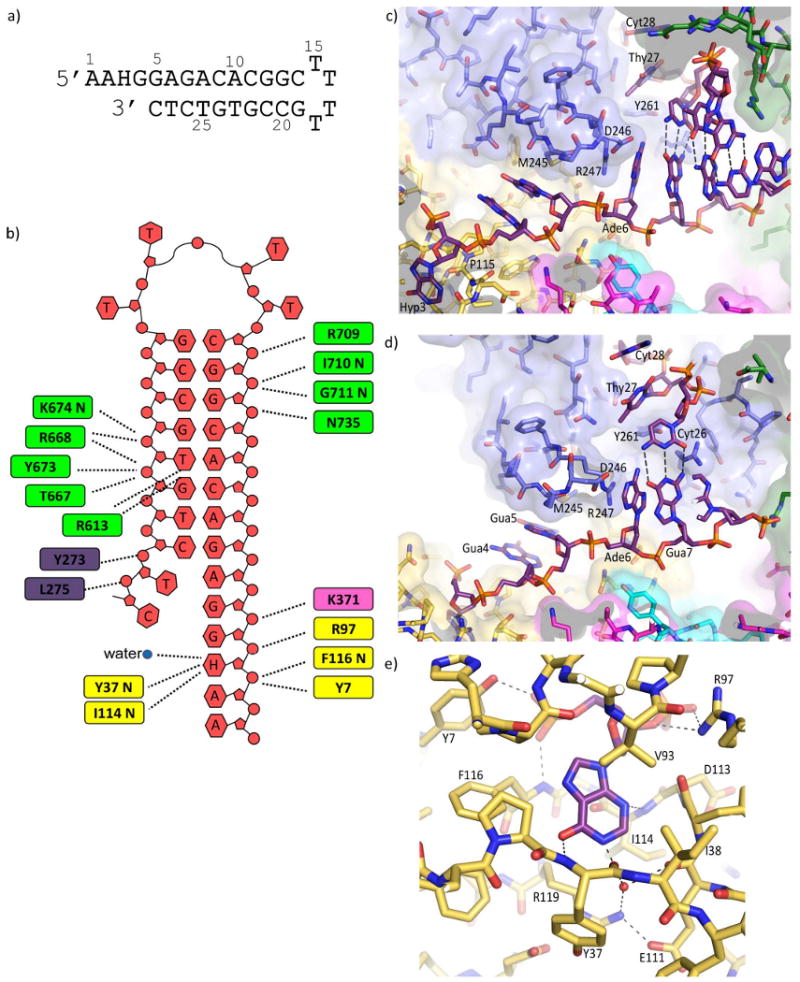
Structure of Tgo-Pol DNA complex. (a) The hypoxanthine-containing DNA used to obtain structural data. The oligodeoxynucleotide folds to produce a primer-template mimic that places hypoxanthine two bases (+2) ahead of the primer-template junction. (b) Summary of the interactions seen between Tgo-Pol and the H+2 primer-template mimic. Although all the bases are shown, A1, A2, and C14 to G19 are not visible in the structure. The extreme 3′ bases (T27 and C28) become single stranded when bound to the polymerase. The amino acids contacting the DNA are color coded according to polymerase sub-domain: thumb, green; palm, pink; amino-terminal, yellow; exonuclease, blue. The interactions made by the β-hairpin to G5 and A6 are not shown. (c and d) Two views illustrating key features of the interactions of the H+2 primer-template mimic with Tgo-Pol. Hypoxanthine (Hyp 3) is bound in the anti-conformation. G4 and G5 are sandwiched between Pro 115 and M245, R247 stacks against A6 and D246 appears to hydrogen bond with this base. As a consequence DNA denaturation occurs with the primer bases T27 and C28 becoming single stranded and well separated from their complementary template bases, G5 and A6. T27 and C28 are located in the editing channel and Y261, which is positioned between T27 and A6, hinders re-annealing. Polymerase amino acids are color coded by sub-domain as for figure 2b. (e) Contacts between Tgo-Pol and hypoxanthine. Three protein-base hydrogen bonds are seen: Tyr 37 (backbone)-O6; Ile 116 (backbone)-N3; Glu 111/R119-N1 (water-mediated). The side chains of E111 and Arg 119 form a salt bridge. The interactions of Tyr 7 and Arg 97 with the flanking phosphates (see also figure 3a) and the stacking of Val 93 with hypoxanthine are also shown.
