Figure 1.
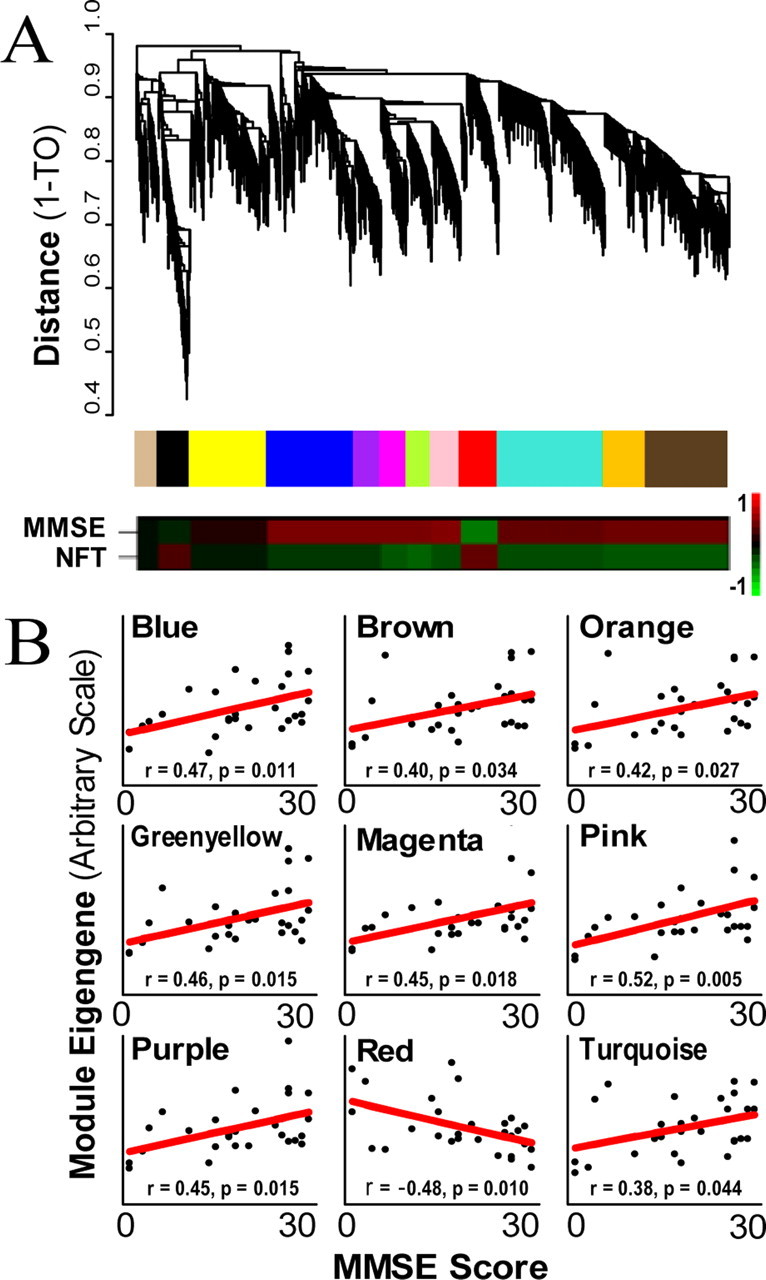
Clustering by topological overlap reveals modules of genes that are characterized by distinct expression patterns. A, Top trace, Cluster dendrogram of genes in the unsupervised AD study groups genes into distinct modules. The y-axis corresponds to distance determined by the extent of topological overlap (1-TO). Dynamic tree cutting was used to identify the most parsimonious module definitions (Materials and Methods), generally dividing modules at significant branch points in the dendrogram. Middle trace, The genes in each of the 12 modules are color-coded. Bottom trace, Heat maps corresponding to the correlation between each ME and both MMSE score and NFT burden. The color scale bar to the right of the bottom trace represents the Pearson correlation ranging from −1 (green) to 1 (red). B, MMSE score (x-axis) plotted vs module eigengene (y-axis) for all nine modules that are significantly correlated with MMSE score. Each point represents a single subject and the line is the line of best fit determined by linear regression across subjects. Only the red module shows negative correlation, indicating an increase in gene expression with AD.
