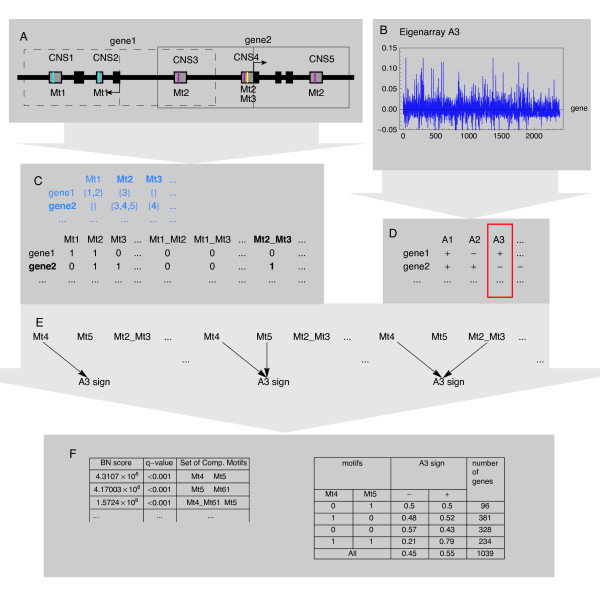
Figure 4.
Bayesian network model of fragmented cis-regulatory regions (A, C) Sequence preprocessing consists of extracting instances of composite motifs i.e. sets of (up to three motifs) in the same conserved non-coding sequence (CNS), from the flanks of transcription start sites of all human-rat orthologous genes. (B, D) Expression data preprocessing consists of SVD, followed by discretization of expression into up- and down-regulation in the subspace of a particular conserved eigensystem - based on the sign of its loading. (C, D) Composite motifs and expression data are combined in one dataset, in which the data records correspond to genes. (E) This dataset becomes an input for our Bayesian networks (BN) learning algorithm, which identifies sets of composite motifs most associated with the sign of loadings of a given eigensystem. (F) The final output consists of a ranking of such sets, with conditional probability distributions representing their impact on a given eigensystem.
BN learning was performed independently for each of the eigensystems: A2, A3, M2, M3; on the data for all the genes in the respective dataset. Eigensystem A3 is shown as an example.