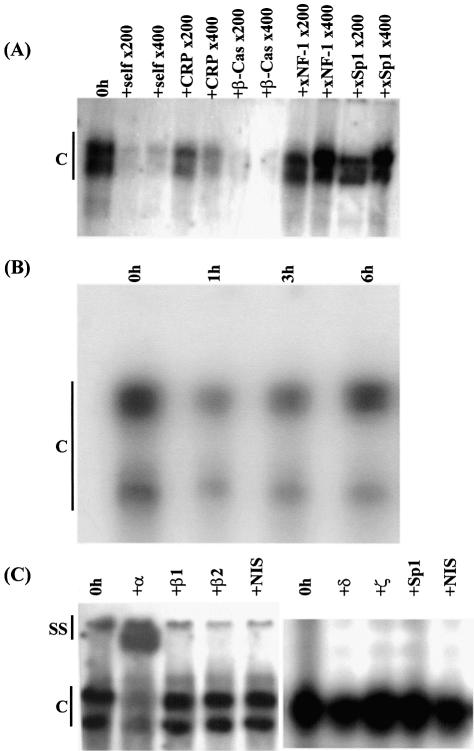
Figure 5.
Analysis of the binding of the C/EBPs to the recognition sequence in the C/EBPα gene promoter. (A) Competition EMSA to verify the specificity of the binding of the C/EBPs to recognition sequence in the xC/EBPα gene promoter. The analysis was carried out using whole cell extracts from untreated Hep3B cells (0 h) and radiolabelled oligonucleotides corresponding to the C/EBP binding site from the xC/EBPα gene promoter. Competitor oligonucleotides were added at 200- and 400-fold molar excess, as indicated, and included the corresponding sequence (self), the C/EBP recognition sequence from the promoters of CRP and β-casein (β-Cas), and the NF-1 and Sp1 sites from the xC/EBPα promoter (xNF-1 and xSp1, respectively). (B) Time-course profile of protein binding to the C/EBP recognition sequence from the mouse C/EBPα gene promoter. EMSAs were carried out using whole cell extracts from Hep3B cells that were treated with IL-6 for the indicated time. (C) Antibody supershift experiments using the C/EBP binding site oligonucleotide. EMSAs were carried out using the C/EBP binding site oligonucleotide from the xC/EBPα gene promoter and whole cell extracts from untreated cells (0 h) in the absence or presence of antisera against C/EBP-α (+α), -β (two different preparations; β1 and β2), -δ (+δ), -ζ (+ζ), Sp1 (+Sp1) and the non-immune serum (NIS). The two closely migrating DNA–protein complexes seen for data shown on the left side of the figure have not separated as efficiently in the case of the gel for the result shown on the right side. Vertical lines labelled C (all panels) and SS (in C only) indicate DNA–protein complexes and antibody–DNA–protein supershift complex, respectively (the free probe has migrated off the gel).