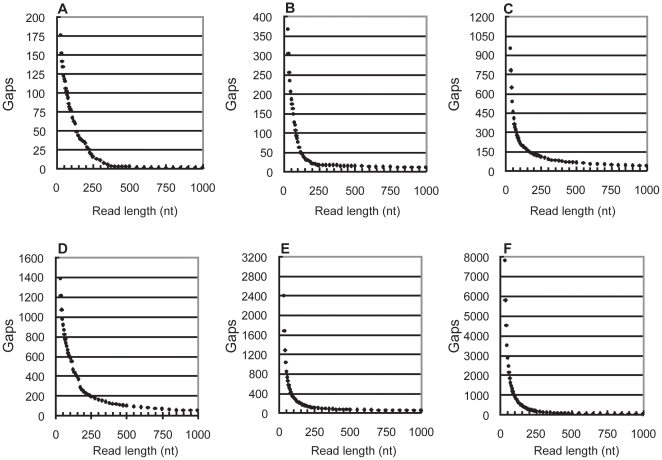
Figure 5. Read length and repeat resolution in 6 genomes.
The algorithm was used to predict the occurrence of repeat-induced gaps in assemblies of six bacterial genomes from a range of read lengths. A raw coverage of 100× was used for all genome/read length pairings. Assembly results were predicted for read lengths at increments between 30–1,000nt. Between 30 and 100nt the increment was 5nt; 100–250nt, 10nt; 250–500nt, 25nt; and 500–1,000nt, 50nt. A) M. genitalium (580 kb), B) H. Influenza (1.8 Mb), C) E. coli (4.6 Mb), D) N. meningitidis (2.3 Mb), E) S. coelicolor (8.7 Mb) and F) S. cellulosum (13.0 Mb).