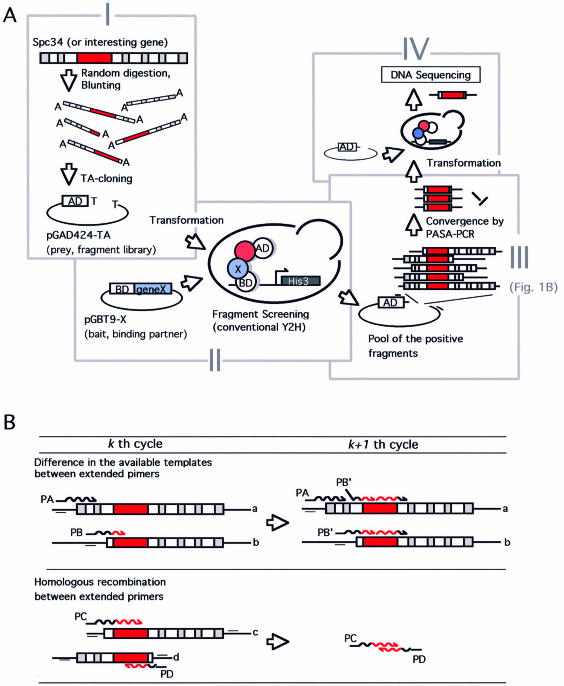
Figure 1.
(A) Schematic representation of the framework for the high-throughput identification of protein interaction domains. (I) Construction of combinatorial ‘endpoints’ library using conventional PCR amplification of the target gene (depicted as Spc34) and DNase I random digestion. The randomly truncated gene fragment was ligated with the two-hybrid vector pGAD424-TA using TA-cloning. The red box in the gene fragment represents the region responsible for the specific interaction. (II) Selection of interaction-positive fragments using conventional yeast two-hybrid screening with a specific binding partner expressing plasmid (depicted as pGBT9-X). (III) Convergence of the selected fragments using PASA-PCR with pGAD424-TA specific primers (represented as small bars on the plasmid). (IV) Polishing of converged fragments. Fragments converged in step III are mixed with EcoRV-digested pGAD424-TA then used for a second yeast transformation. Several clones on the selection plate are subjected to DNA sequence analysis to identify the position of critical endpoint. The region drawn in red represents the protein interaction domain. See text for more details. (B) Convergence of the selected (binding-positive) fragments by PASA-PCR. The mechanism of the preferential amplification of the shortest amplicon is illustrated. (Upper) Preferential amplification based on the difference in effective concentration of templates. PA, primer extended on template with long flanking sequence (template a). PB, primer extended on short template with less flanking sequence (template b). This diagram represents that only template a is available for further extension of PA in the next (k + 1th) cycle, whereas the PB can utilize the both to give further extended primer PB’, resulting in more efficient amplification. (Lower) A homologous recombination between the two primers prematurely extended in kth cycle (PC, primer extended on template c, and PD, the counterpart primer extended on template d) occurs to form a shorter fragment with the novel combination of the endpoints in (k + 1th) cycle. See text for details.