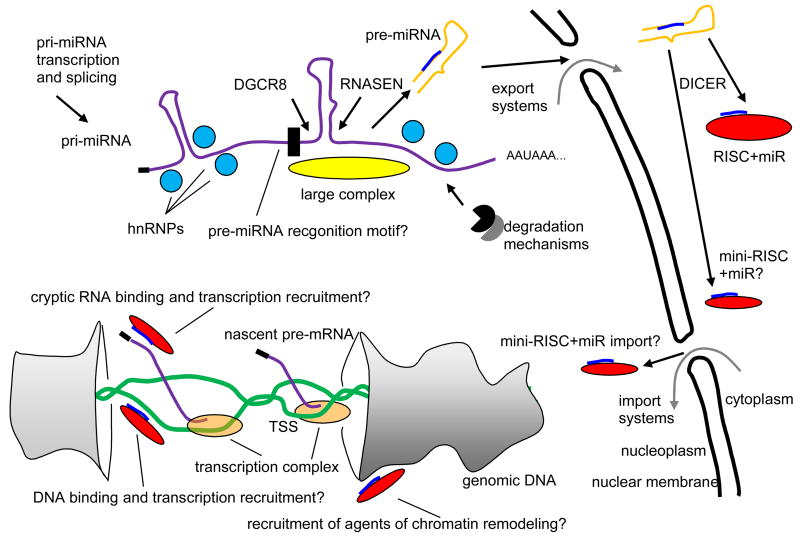
Fig 6.
Mechanisms, largely speculative or surmised, suggested in this paper. A primary miRNA transcript (pri-miRNA), possibly from an intron, is chaperoned into folded RNA (purple) by heterogeneous ribonucleoprotein particles (hnRNPs) (light blue), countering RNA degradation mechanisms. Possibly a large complex (Gregory et al., 2004) (yellow) or a sequential or geometric recognition motif (black) guide recruitment of protein products of genes DGCR8 and RNASEN, yielding excision of a precursor RNA hairpin (pre-miRNA) (orange with blue subsequence that becomes the mature miRNA). The pre-miRNA is exported. Protein product of the DICER gene selects an ssRNA section of the hairpin stem, the mature miRNA, for mounting on an RNA induced silencing complex (RISC) (red). Evidence exits (Ohrt et al., 2008) that a single protein product of AGO2 enables re-import of some miRNAs or other short ssRNAs, possibly as a “mini-RISC.” Back in the nucleus the miRNA might engage open genomic DNA (green) or some cryptic noncoding RNA to enhance or suppress recruitment of transcription complex to the transcription start site (TSS) of a gene. Recruitment of agents of chromatin remodeling could be part of the mechanisms.