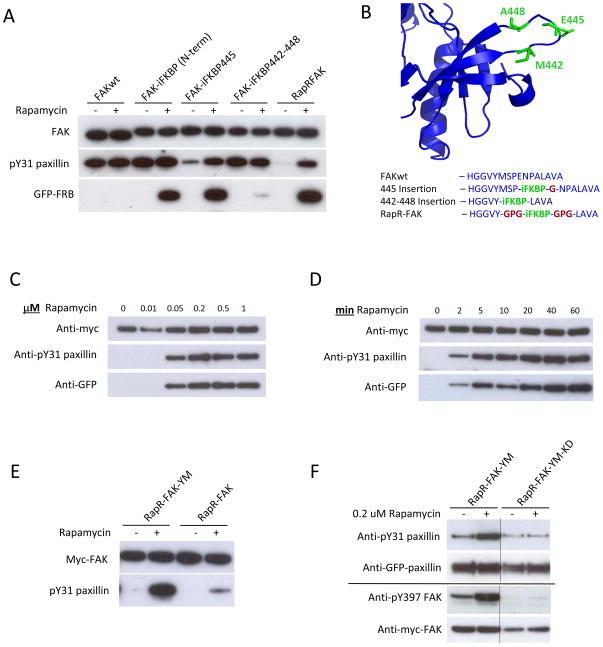
Fig. 2.
Development and biochemical characterization of RapR-FAK. (A) Rapamycin regulation of FAK variants with iFKBP inserted at different positions. HEK293T cells co-expressing myc-tagged FAK constructs and GFP-FRB were treated for one hour with either 200 nMrapamycin or ethanol (solvent control). The activity of immunoprecipitated FAK variants was tested using the N-terminal fragment of paxillin as a substrate. (B) Sites of iFKBP insertion (green) and connecting linkers (red). (C, D) HEK293T cells co-expressing RapR-FAK and FRB were treated with the indicated amount of rapamycin for 1 hour or with 200 nMrapamycin for the indicated period of time. The kinase was immunoprecipitated and its activity tested as described above. (E) FAK Y180A and M183A mutations were introduced to eliminate autoinhibitory interactions, thereby generating RapR-FAK-YM, which was tested as in A. (F) HEK293T cells co-expressing Cherry-FRB, GFP-paxillin and either myc-tagged RapR-FAK-YM or its kinase-inactive mutant (RapR-FAK-YM-KD) were treated with rapamycin or ethanol (solvent control) for 1 hour. GFP-paxillin was immunoprecipitated and its phosphorylation was assessed using anti-phospho-Tyr31 antibody. Autophosphorylation of FAK on Tyr397 was analyzed using total cell lysate.