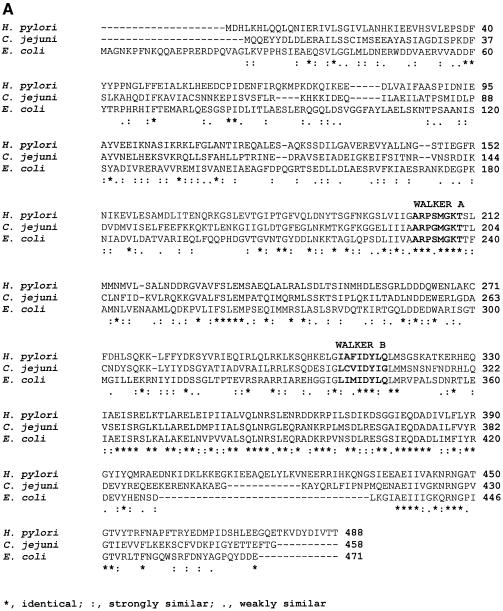
Figure 1.
Primary sequence analysis of HpDnaB and in vivo expression. (A) Protein sequence alignment of homologous DnaB proteins. Comparison of the H.pylori, C.jejuni and E.coli DnaB primary structures. The amino acid sequences of the respective proteins were aligned by CLUSTALW multiple alignment program. ‘*’ indicates the residues which are identical, ‘:’ indicates the residues which are strongly similar and ‘.’ indicates the residues which are weakly similar. Nucleotide binding (Walker A) and hydrolysis (Walker B) motifs are also shown. (B) In vivo expression of DnaB in H.pylori. Western blot of H.pylori and E.coli cell extract using a polyclonal antisera against DnaB (E.coli). Lane 1 contains 1.0 µg of purified E.coli DnaB, lanes 2 and 3 contain cell extract of H.pylori (5.0 and 10.0 µg, respectively), lanes 4 and 5 contain cell extract of E.coli (5.0 and 10.0 µg, respectively) and lane 6 contains 100 ng of purified GST-tagged DnaB of H.pylori.