Figure 1.
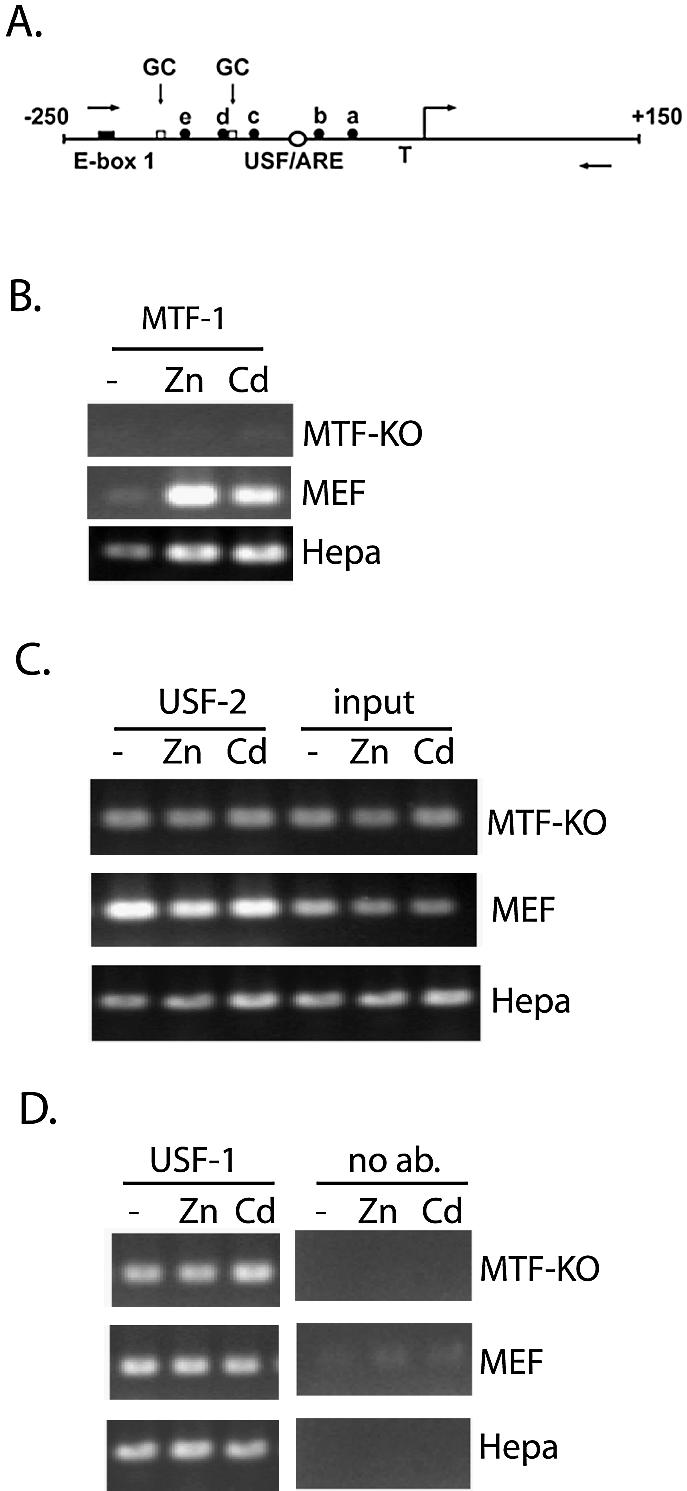
Interactions of USF-1 and -2 in vivo with the MT-I promoter are metal-independent and MTF-1-independent. (A) Schematic of the proximal promoter of the MT-I gene. The nucleotide sequence (bp) is relative to the transcriptional start site (right angle arrow). The relative locations of the USF/ARE (centered at –95 bp), which contains a sequence homologous to an AP-1 consensus binding site, E-box 1 (centered at –220 bp), the multiple MREs (a–e), and the TATA box (T) are illustrated. Two GC-boxes (one centered at –183 bp; the other immediately downstream of MRE-d) represent putative binding sites for Sp transcription factors. The locations of PCR primers used in the ChIP assay are indicated using forward (sense) and reverse (antisense) arrows. (B–D) Chromatin immunoprecipitation (ChIP) assay of the proximal MT-I promoter. Chromatin was isolated from Hepa cells, wild-type mouse embryonic fibroblasts (MEF) or fibroblasts lacking expression of MTF-1 (MTF-KO) that remained untreated (–), or were treated with 100 µM zinc (Zn) or 10 µM cadmium (Cd) for 3 h. ChIP was carried out using antibodies against MTF-1 (B), USF-2 (C), or USF-1 (D), as described in the text. Shown in (C), a portion of the chromatin (0.5% of that used for all of these immunoprecipitations) was purified and analyzed by PCR prior to the immunoprecipitation step (input). Fold-induction for metal-induced binding of transcription factors to the chromatin, enumerated in the text, was calculated from triplicate PCRs after normalizing antibody-specific PCR products to input PCR products.
