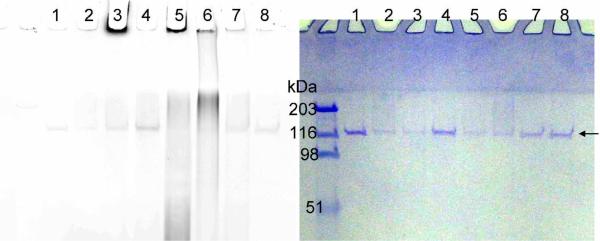
Figure 3.
Labeling of PARP-1 auto-(ADP-ribosyl)ation with 6- or 8-alkyne-NAD. The panel on the left shows the image of Rhodamine fluorescence and the panel on the right is the same gel stained with Coomassie blue. The arrow points at the position of unmodified PARP-1. All lanes contain PARP-1 (0.15 μM) and all lanes except 7 and 8 contain ssDNA (0.25 μg/μL). Lane 1, control without NAD; lane 2, control with normal NAD (100 μM); lane 3, 6-alkyne-NAD (100 μM); lane 4, 8-alkyne-NAD (100 μM); lane 5, 6-alkyne-NAD (100 μM) and NAD (100 μM); lane 6, 8-alkyne-NAD (100 μM) and NAD (100 μM); lane 7, control with 6-alkyne-NAD (100 μM), NAD (100 μM), and no ssDNA; lane 8, control with 8-alkyne-NAD (100 μM), NAD (100 μM), and no ssDNA. The fluorescence signal below 116 kDa in lane 5 and 6 is likely due to the hydrolysis of poly(ADP-ribose) chain resulting in poly(ADP-ribose) polymers that are not covalently bound to proteins. The linkage between different ADP-ribose units is a relatively labile ester bond.