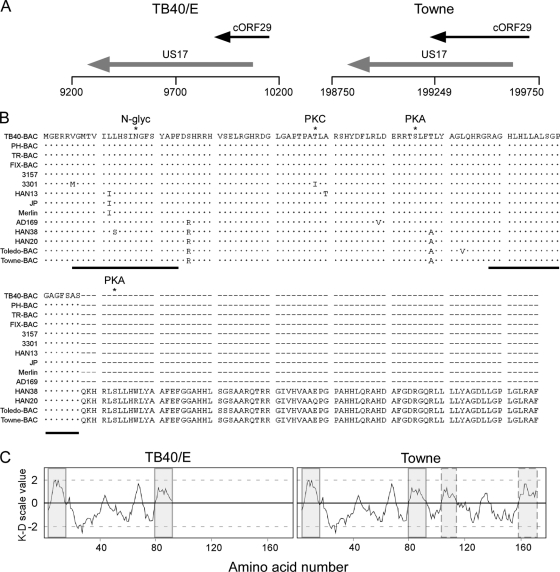
FIG. 1.
Genomic location of the c-ORF29 gene and in silico analysis of the RASCAL amino acid sequence. (A) Schematic map of the TB40-BAC4 (GenBank accession no. EF999921) and of the Towne-BAC (accession no. AY315197) genomic regions corresponding to nucleotides 9200 to 10200 (TB40/E) and 198750 to 199750 (Towne). The black horizontal line depicts the viral genome, with vertical lines positioned every 500 nucleotides. ORFs are represented by arrows pointing in the direction of transcription. (B) ClustalW2 alignment of RASCAL amino acid sequences from 14 human CMV strains. Viral genome accession numbers are as follows: TB40-BAC, EF999921; PH-BAC, AC146904; TR-BAC, AC146906; FIX-BAC, AC146907; 3157, GQ221974; 3301, GQ466044; HAN13, GQ221973; JP, GQ221975; Merlin, AY44689; AD169, X17403; HAN38, GQ396662; HAN20, GQ396663; Toledo-BAC, AC146905; and Towne-BAC, AC146851. Dots and dashes indicate identical and absent amino acids, respectively. The asterisks mark the specific amino acid predicted to be phosphorylated by PKA or PKC or to be N glycosylated (N-glyc). The black lines underscore the putative TMDs. (C) Kyte-Doolittle (K-D) hydropathy plot of RASCALTB40/E and RASCALTowne amino acid sequences, performed using a window size of 9 aa. The shaded boxes include the stretch of amino acids corresponding to the putative TMDs, while the boxes with the dashed borders include the two additional hydrophobic regions of more than 17 aa detected in RASCALTowne.