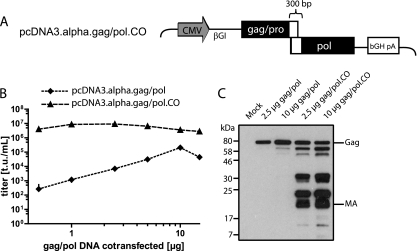
FIG. 3.
Codon optimization of the gag/pol helper plasmid. (A) Schematic depiction of pcDNA3.alpha.gag/pol.CO, showing the cytomegalovirus promoter (CMV), the β-globin intron (βGI), and the bovine growth hormone polyadenylation signal (bGH pA). Solid rectangles represent areas of codon optimization, while braces indicate sequence that has been exempted from the codon optimization of pcDNA3.alpha.gag/pol. (B) Dilution series of gag/pol helper plasmids. Five micrograms of pAlpha.SIN.SF.EGFP.wPRE, 1.5 μg of the VSVg envelope expression plasmid, and different amounts of either pcDNA3.alpha.gag/pol or its codon-optimized counterpart were cotransfected into 293T cells. Viral supernatants were titrated on HT1080 cells. (C) Western blot of 293T cells. Cells were cotransfected with an ecotropic envelope expression plasmid, pAlpha.SIN.SF.EGFP.wPRE, and different amounts of either pcDNA3.alpha.gag/pol or its codon-optimized counterpart (2.5 or 10 μg, as indicated). Viral proteins were detected by using antibodies against the alpharetroviral matrix (MA) protein. Molecular mass standards are given on the left, while Gag and MA are indicated on the right.