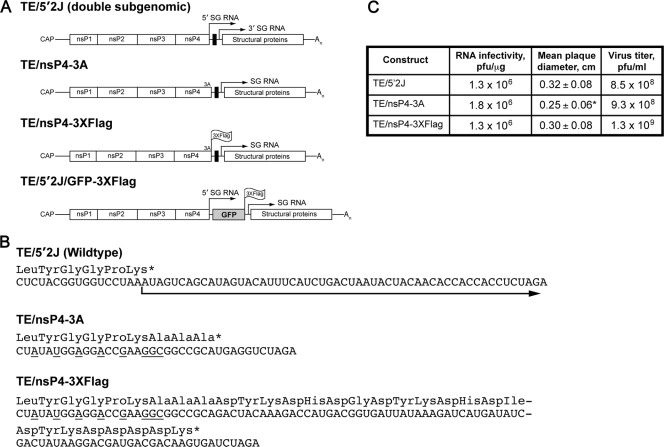
FIG. 1.
Characteristics of viruses used in the present study. (A) Schematic diagrams of the viruses utilized in this study are shown. TE/5′2J expresses two subgenomic (SG) RNAs, indicated by arrows above the diagram; the 5′ SG RNA express foreign genes inserted using restriction sites (black box) between the two SG transcriptional start sites, while the 3′ SG RNA encodes the viral structural proteins. An indicates the poly(A) tail. Three alanine residues expressed by SINV TE/nsP3-3A are indicated at the carboxyl terminus of nsP4, the viral RNA-dependent RNA polymerase. The location of the 3×Flag tag at the carboxyl termini of nsP4 and GFP in SINV TE/nsP4-3×Flag and TE/5′2J/GFP-3×Flag, respectively, is indicated. (B) The RNA sequences of the parental (TE/5′2J) and derived viruses are shown, with the amino acid sequence shown above. An asterisk indicates a stop codon. The arrow indicates the SG RNA derived from the 5′ SG promoter. Nucleotides mutated to disrupt SG promoter function are underlined. (C) Comparison of the infectivity of the viral constructs. BHK-21 cells were electroporated with 1 μg of each RNA in duplicate (or triplicate, TE/nsP4-3×Flag), and the number of infectious centers was determined. Mean plaque diameter, as assessed by measurement of 30 to 40 plaques for each virus, is shown, with the standard deviation indicated. The asterisk indicates a statistically different mean diameter from the TE/5′2J parental virus (P = 0.0015, unpaired t test). The remaining electroporated cells were placed in culture and 1 day later the medium overlying the cells was removed and pooled to generate a virus stock. The titer of infectious virus, determined by standard plaque assay, is indicated.