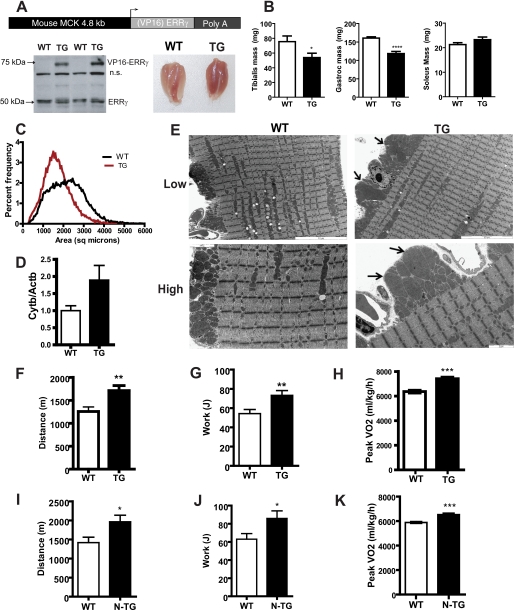
FIGURE 2.
Mice expressing ERRγ in skeletal muscle have “red” muscle, larger mitochondria, and improved oxidative capacity. A, schematic representing the structure of the transgene used to generate the ERRγ transgenic mice. The mouse muscle creatine kinase was used to drive the expression of native ERRγ or VP16ERRγ fusion protein. As shown in the lower left panel, in the VP16ERRγ transgenics, the protein was expressed in the skeletal muscle as observed by Western blot using ERRγ antibody. The endogenous ERRγ band at 50 kDa is indicated. The lower right panel shows a picture of the gastrocnemius-soleus muscle from the wild type (WT) and VP16ERRγ transgenic (TG) mice. n.s., not significant. B, individual muscle weights from the VP16ERRγ (TG) and WT mice (n = 12 mice per group). C, plot showing the size distribution of individual muscle fibers in VP16ERRγ transgenic (red) versus control littermate muscle (WT, black) (n = 5–6 mice per group). D, mitochondrial DNA content from gastrocnemius muscle from WT and VP16ERRγ TG mice (n = 8 mice per group). E, electron micrographs for gastrocnemius muscle from WT (left panels) and VP16ERRγ TG (right panels) mice. The upper panels represent a lower magnification (×5310), and the bottom panels are images taken at a higher magnification (×12,600). The scale for each image is embedded within the image. In the TG images, arrows point to enlarged mitochondria. Images shown are representative from n = 4 per genotype. Exercise capacity determined on a treadmill for the both VP16ERRγ (designated as TG, F–H) and ERRγ (designated as N-TG, I–K) transgenic mice along with their corresponding control WT mice. Data presented are distance run (F and I) and work performed (G and J) (n = 12 mice per group for VP16ERRγ strain and n = 10 for ERRγ strain). Peak oxidative capacity was assessed in WT and TG mice (H) and WT and N-TG mice (K), on a treadmill (n = 10 mice per group for both strains) under a peak VO2 protocol. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.005.