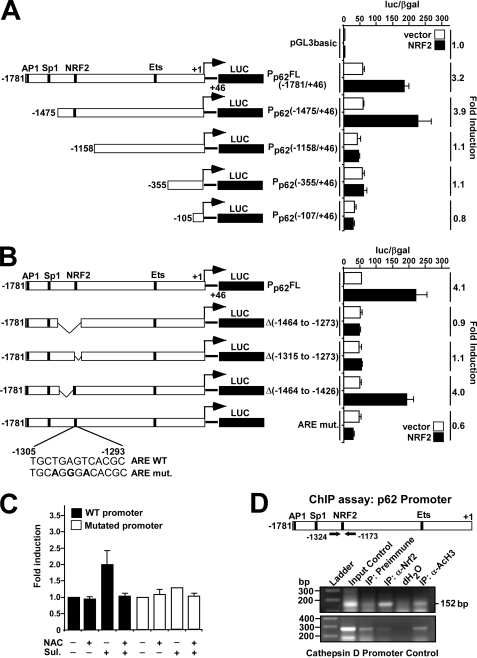
FIGURE 1.
Mapping of an NRF2 binding site in the p62 promoter/enhancer. A and B, reporter gene assays were performed using wild-type (−1781/+46) or the indicated deleted or mutated p62 promoter/enhancer constructs. HEK293 cells were co-transfected with an empty vector (pcDNA3.1) (100 ng) or a plasmid encoding murine Nrf2 (pcDNA3.1-V5-mNrf2) (100 ng) together with the indicated p62 promoter constructs (60 ng). Cells were harvested 24 h after transfection. The relative promoter activities are expressed as the ratio between measured luciferase and β-galactosidase activities. The data shown are the mean activities obtained in one experiment performed in triplicate, and are representative of three or more independent experiments. For each p62 promoter construct, NRF2-mediated fold-activation is shown to the right. C, HEK293 cells co-transfected with wild-type or mutated p62 promoter constructs were analyzed as in A. The cells were treated for 20 h with sulforaphane (Sul, 15 μm) or N-acetylcysteine (NAC, 5 mm) as indicated. The relative activity of the wild-type promoter was set to one. The mean -fold activation obtained in three independent experiments performed in triplicate is presented. D, ChIP assays show that NRF2 can associate with the p62 promoter. Extracts from HeLa cells (1.5 × 107 cells per antibody) were immunoprecipitated with preimmune serum, polyclonal anti-NRF2 antibody, or anti-acetylated histone H3 antibody as a positive control. Input control (1:50) was also included. PCR analyses of the immunoprecipitated chromatin were carried out using primers flanking the ARE (position −1324 and −1173, respectively) (upper panel). PCR analyses of the precipitated chromatin using primers aligning to position −3351 and −3069 of the cathepsin D promoter were used as a control.