cdc15-2 cells failed to coordinate spindle elongation and kinetochore capture during recovery from Noc treatment. This was due to a failure in the cdc15-2 cells to accumulate Pds1p as well as in wild-type cells, indicating that timely progression through the cell cycle and proper expression of PDS1 is important in the presence of spindle damage.
Abstract
During normal metaphase in Saccharomyces cerevisiae, chromosomes are captured at the kinetochores by microtubules emanating from the spindle pole bodies at opposite poles of the dividing cell. The balance of forces between the cohesins holding the replicated chromosomes together and the pulling force from the microtubules at the kinetochores result in the biorientation of the sister chromatids before chromosome segregation. The absence of kinetochore–microtubule interactions or loss of cohesion between the sister chromatids triggers the spindle checkpoint which arrests cells in metaphase. We report here that an MEN mutant, cdc15-2, though competent in activating the spindle assembly checkpoint when exposed to Noc, mis-segregated chromosomes during recovery from spindle checkpoint activation. cdc15-2 cells arrested in Noc, although their Pds1p levels did not accumulate as well as in wild-type cells. Genetic analysis indicated that Pds1p levels are lower in a mad2Δ cdc15-2 and bub2Δ cdc15-2 double mutants compared with the single mutants. Chromosome mis-segregation in the mutant was due to premature spindle elongation in the presence of unattached chromosomes, likely through loss of proper control on spindle midzone protein Slk19p and kinesin protein, Cin8p. Our data indicate that a slower rate of transition through the cell division cycle can result in an inadequate level of Pds1p accumulation that can compromise recovery from spindle assembly checkpoint activation.
INTRODUCTION
The timely execution of chromosome segregation such that it precedes mitotic exit and cytokinesis is important for the dividing cells, as premature exit from mitosis and cytokinesis before sister chromatid separation between mother and daughter cells would possibly result in improper nuclear division and loss of genomic stability. In the budding yeast, sister chromatid segregation is normally prevented by the inhibitor of anaphase, Pds1p (securin), that acts to inhibit the activity of Esp1p (separase) from cleaving the cohesins that keep the sister chromatids together (Nasmyth, 2002). The destruction of securin depends on an E3 ubiqutin-ligase known as the anaphase-promoting complex (APC) and an activator of the APC known as Cdc20p (reviewed in Thornton and Toczyski, 2006). Exit from mitosis requires the destruction of the mitotic cyclin, Clb2 (Morgan, 1999) and depends on Cdc20p and a homologue of Cdc20p known as Hct1p (Visintin et al., 1997). The activation of Hct1p itself is under the regulation of a pathway known as the mitotic exit network (MEN). The MEN components include Tem1p (a GTPase); Lte1p (a GTP/GDP exchange factor); Cdc15p, Cdc5p, Dbf2p, Dbf20p (Ser/Thr kinases); and Cdc14p (a phosphatase; reviewed in Lee et al., 2001; Sullivan and Morgan, 2007). The coordinated activation of APCCdc20p and APCHct1p (Bardin and Amon, 2001; Yeong et al., 2002; Simanis, 2003) helps trigger the destruction of securin and Clb2p such that cells separate sister chromatids before exit from mitosis during normal progression through mitosis.
During mitosis, the biorientation of chromosomes ensures accurate sister chromatid partitioning (reviewed in Tanaka et al., 2005; Musacchio and Salmon, 2007). Biorientation of sister chromatids occurs during metaphase when microtubules emanating from opposite spindle pole bodies (SPBs) attach themselves to the kinetochores on a pair of chromatids (Westermann et al., 2007) held together by cohesins (Uhlmann, 2004). The force of the microtubules pulling at the kinetochores coupled with the cohesins keeping the sister chromatids generates a tension in the chromosome-microtubule structure (Tanaka et al., 2005; Musacchio and Salmon, 2007). In metaphase when there is a loss of kinetochore–microtubule attachment or loss of sister cohesion, either of which results in the reduction of tension in the structure, the spindle checkpoint is activated. The checkpoint imposes a delay on cell cycle progression such that cells arrest in metaphase. On restoration of the spindle attachment to the kinetochores, the spindle checkpoint switches off, and the cells continue along their pathway toward segregating sister chromatids and mitotic exit.
The spindle checkpoint includes components such as Mad1p–Mad3p, Bub1p, Bub3p, Ipl1p, and Mps1p (Musacchio and Salmon, 2007). These components serve to sequester Cdc20p from the APC during checkpoint activation, thereby preventing the ubiquitination and consequently, destruction of securin. As a result, separase is held in check and sister chromatids separation is prevented. The MEN pathway is the target for the spindle position checkpoint. The spindle position checkpoint depends on Bub2p and monitors the orientation and position of the spindle apparatus. In the event that the spindle apparatus is mis-oriented, Bub2p acts to prevent that activation of the MEN pathway to delay mitotic exit until the spindle apparatus is properly oriented (Lew and Burke, 2003).
In our present study, we report that cdc15-2 cells mis-segregated sister chromatids during recovery from the spindle assembly checkpoint activation even though the cells were able to arrest in nocodazole (Noc). We provide data showing that this was due to the untimely elongation of the mitotic spindles, as the cdc15-2 mutant cells failed to accumulate sufficiently high levels of Pds1p during exposure to Noc. Our data support the notion that a lack of proper accumulation of Pds1p during a delayed cell cycle progression while cells are exposed to Noc can affect chromosome segregation during recovery from checkpoint activation. Therefore, in addition to a functional spindle assembly checkpoint, a timely transit through the cell division cycle during response to spindle damage helps ensure proper inheritance of chromosomes.
MATERIALS AND METHODS
Strains and Plasmids
A combination of standard molecular biology and molecular genetic techniques such as PCR-based tagging of endogenous genes and tetrad dissection were used to construct plasmids and strains with various genotypes (Table 1). The plasmids for the green fluorescent protein (GFP), Redstar2, and Myc cassettes were obtained from EUROSCARF (Frankfurt, Germany). Details of the primers used for the strain constructions will be provided upon request.
Table 1.
List of strains
| Name | Genotype | Source |
|---|---|---|
| US1363 | MAT a leu2 his3 trp1 ura2 bar1S | Surana lab |
| US1318 | MAT a cdc15-2 leu2 his3 trp1 ura2 bar1 | Surana lab |
| FM689 | Mat a MYO1-Redstar2∷NAT IPL1-GFP∷KAN | This study |
| FM700 | Mat a cdc15-2 MYO1-Redstar2∷NAT IPL1-GFP∷KAN | This study |
| FM695 | Mat a MYO1-Redstar2∷NAT SLI15-GFP∷HIS3 | This study |
| FM716 | Mat a cdc15-2 MYO1-Redstar2∷NAT SLI15-GFP∷HIS3 | This study |
| FM771 | MAT a GFP-TUB1-URA3∷TUB1 MYO1-Redstar2∷NAT | This study |
| FM756 | MAT a cdc15-2 GFP-TUB1-URA3∷TUB1 MYO1-Redstar2∷NAT | This study |
| FM990 | MAT a leu2∷LEU2 tetR-GFP 1.4 kb left ofCEN5∷tetO2X112∷HIS3 MYO1-Redstar2∷NATaSPC29-RFP∷HYGRO | This study |
| FM991 | MAT a cdc15-2 leu2∷LEU2 tetR-GFP 1.4 kb left of CEN5∷tetO2X112∷HIS3 MYO1-Redstar2∷NAT SPC29-RFP∷HYGROb | This study |
| FM1044 | Mat a cdc20Δ∷LEU2 GAL-CDC20∷TRP SPC29-RFP∷HYG BAR NDC80-GFP∷ HIS3MX6 | This study |
| FM1033 | Mat a cdc15-2 cdc20Δ∷LEU2 GAL-CDC20∷TRP SPC29-RFP∷HYG BAR NDC80-GFP∷ HIS3MX6 | This study |
| FM1051 | Mat a NDC80-Redstar2∷NAT MAD2-GFP∷ HIS3MX6 | This study |
| FM1041 | Mat a cdc15-2 NDC80-Redstar2∷NAT MAD2-GFP∷ HIS3MX6 | This study |
| FM1058 | Mat a cdc20Δ∷LEU2 GAL-CDC20∷TRP leu2∷LEU2 tetR-GFP 1.4 kb left of CEN5∷tetO2X112∷HIS3 SPC29-RFP∷HYGRO | This study |
| FM1059 | Mat a cdc15-2 cdc20Δ∷LEU2 GAL-CDC20∷TRP leu2∷LEU2 tetR-GFP 1.4 kb left of CEN5∷tetO2X112∷HIS3 SPC29-RFP∷HYGRO | This study |
| FM1068 | MAT a PDS1-9MYC∷HIS3MX6 bar1 | This study |
| FM1117 | MAT cdc15-2 PDS1-9MYC∷HIS3MX6 bar1 | This study |
| FM1294 | MAT a mad2Δ PDS1-9MYC∷HIS3MX6 bar1 | This study |
| FM1296 | MAT a cdc15-2 mad2Δ PDS1-9MYC∷HIS3MX6 bar1 | This study |
| FM1545 | MAT a bub2Δ PDS1-9MYC∷HIS3MX6 bar1 | This study |
| FM1587 | MAT a cdc15-2 bub2D PDS1-9MYC∷HIS3MX6 bar1 | This study |
| FM1364 | MAT pMET-CDC20∷TRP NDC80-GFP∷HIS 1.4 kb left of CEN5∷tetO2X112∷HIS3 tetR-mRFP on LEU, CEN plasmid | This study |
| FM1347 | MAT a cdc15-2 pMET-CDC20∷TRP NDC80-GFP∷HIS 1.4kb left of CEN5∷tetO2X112∷HIS3 tetR-mRFP on LEU, CEN plasmid | This study |
| FM1549 | MAT a CIN8-GFP∷URA MYO1-Redstar∷NAT SPC29-RFP∷HYGRO | This study |
| FM1460 | MAT a cdc15-2 CIN8-GFP∷URA MYO1-Redstar∷NAT SPC29-RFP∷HYGRO | This study |
| FM1543 | MAT a slk19Δ∷KANMX4 CIN8-GFP∷URA SPC29-RFP∷HYGRO | This study |
| FM1526 | MAT a cdc15-2 slk19Δ∷KANMX4 CIN8-GFP∷URA SPC29-RFP∷HYGRO | This study |
| FM1665 | MAT a cdc15-2 leu2∷LEU2 tetR-GFP 1.4 kb left of CEN5∷tetO2X112∷HIS3 MYO1-Redstar2∷NAT SPC29-RFP∷HYGRO MET-PDS1myc∷URA | This study |
| FM1677 | MAT a MET-PDS1-myc∷LEU SPC29-RFP∷HYG | This study |
| FM1678 | MAT a cdc15-2 MET-PDS1-myc∷LEU SPC29-RFP∷HYG | This study |
| FM1689 | MAT a bar1 PDS1-GFP∷KAN SPC29-RFP∷HYG | This study |
| FM1687 | MAT a cdc15-2 bar1 PDS1-GFP∷KAN SPC29-RFP∷HYG | This study |
| FM | Mat a cdc20Δ∷LEU2 GAL-CDC20∷TRP leu2∷LEU2 tetR-GFP 1.4 kb left of CEN5∷tetO2X112∷HIS3 SPC29-RFP∷HYGRO Pds1-HA∷NAT | This study |
| FM1699 | Mat alpha cdc15-2 cdc20Δ∷LEU2 GAL-CDC20∷TRP leu2∷LEU2 tetR-GFP 1.4 kb left of CEN5∷tetO2X112∷HIS3 SPC29-RFP∷HYGRO Pds1-HA∷NAT | This study |
a NAT, Nourseothricin.
b HYGRO, Hygromycin B.
Yeast Culture Reagents
Wild-type haploid W303 strain was used in this study. Cells were routinely grown in yeast-extract peptone (YP) or selective medium supplemented with 2% dextrose (D) at 24°C. For experiments requiring galactose (Gal) induction, cells were grown in YP supplemented with 2% raffinose (Raff) followed by addition of Gal to a final concentration of 2% unless otherwise stated.
Synchronization Procedures
For experiments requiring synchronized cultures, exponential phase cells were diluted to 107 cells/ml in growth medium at 24°C. For G1 arrest, cells were treated with α-factor at 0.4 μg/ml for 3 h. After the cells were arrested, they were washed by filtration and resuspended in media at the required conditions as described in the various sections. For a typical Noc arrest, cells were arrested with 7.5 μg/ml Noc for 2.5 h at 24°C followed by the further addition of 7.5 μg/ml for another 2.5 h at 32°C. The drug was washed off by centrifugation of the cells. Cells were then released and sampled at intervals as described in the relevant sections. Each experiment was performed three times. The number of cells counted for each typical experiment shown is stated in the relevant sections. Graphs shown are representative of a typical experiment out of three experiments.
Western Blot Analysis
Western blot analyses were performed as previously described (Yeong et al., 2000). Anti-Cdc28p antibodies, anti-Clb2 antibodies, anti-myc and anti-hemagglutinin (HA; Santa Cruz Biotechnology, Santa Cruz, CA) were used at a 1:1000–1:2000 dilution. An enhanced chemiluminescence kit (Pierce Chemical, Rockford, IL) was used according to the manufacturer's recommendations.
Fluorescence Microscopy
Cells carrying GFP and red fluorescent protein (RFP) fusions were washed in dH2O and observed directly without fixation after washing. Samples were observed using an Olympus IX81 epifluorescence microscope (Melville, NY), with a 60× oil lens (NA 1.4) and a 1.5× Opitvar. Filter sets for the fluorescence proteins were from Omega Optical (Brattleboro, VT) and Semrock (Rochester, NY), and images were captured using CoolSnapHQ CCD (Photometrics, Tucson, AZ). Image acquisition was controlled by Metamorph (Molecular Devices, Sunnyvale, CA). Typically, for single plane wide-field images, the exposure time for GFP was ∼0.8–1 s and RFP was ∼0.6–0.8 s. For spinning disk confocal images, typically exposure time for the spinning disk images was ∼0.3–0.8 s per plane. Eleven optical Z-sections at 0.3- or 0.5-μm intervals were obtained, and the images shown are maximal projections of the Z-sections.
Time-Lapse Microscopy
For time-lapse microscopy, cells released from a Noc arrest were resuspended in complete synthetic media (SC) containing either glucose (Glu) or Raff or Raff and Gal and mounted onto 5% agarose pads on slides. Temperature control was achieved using the Bioptechs lens heater (Butler, PA) calibrated at 30.6°C. Spinning disk time-lapse images were obtained using an Olympus IX81 ZDC microscope, with a 60× oil lens (NA 1.4) and a 1.5× Opitvar. An argon-krypton laser source was used for the excitation of the samples. Filter sets for the fluorescence proteins were from Omega and Semrock, and images were captured using the Photometrics 512EM-CCD attached behind the Yokogawa CSU22 (Tokyo, Japan) connected to the microscope. Where compatible, GFP and RFP images were capture simultaneously using via a Dual-View image-splitter (Optical Insights, Tucson, AZ). Image acquisition was controlled by Metamorph. Images were captured at the time intervals as indicated in the relevant sections. Typically, the exposure time for the spinning disk images was ∼0.3–0.8 s per plane. Eleven optical Z-sections at 0.3- or 0.5-μm intervals were obtained for each time point. Images shown are maximal projection of the Z-stacks. Image J (http://rsb.info.nih.gov/ij/) and Adobe Photoshop (San Jose, CA) were used for the production of the montages and figures. The number of cells counted for the time-lapse experiments shown was taken together from at least three different experiments.
RESULTS
cdc15-2 Cells Mis-Segregate Chromosome V during Recovery from Noc Treatment
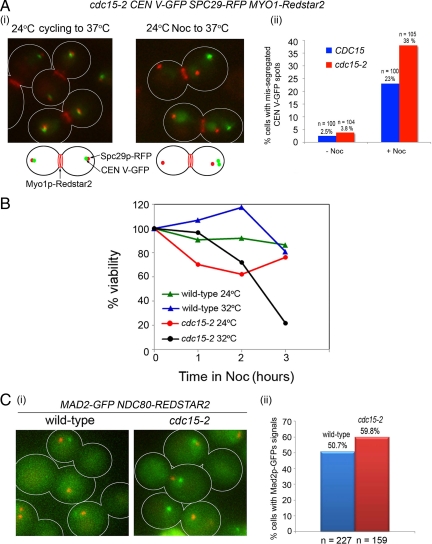
We previously noted that cdc15-2 tetR-GFP CEN5∷tetO2X112 SPC29-RFP MYO1-Redstar2 cells, when arrested in the microtubule-depolymerizing drug Noc at 24°C and released into the restrictive temperature of 37°C, mis-segregated CEN V-GFP (Figure 1Ai, right) in 38.0% of the cells (Figure 1Aii). The chromosome mis-segregation appeared related to the Noc treatment, as cdc15-2 cells shifted directly to 37°C from 24°C without prior exposure to Noc (Figure 1Ai, left) only showed 3.8% (Figure 1Aii) CEN V-GFP mis-segregation. Wild-type cells treated similarly consistently showed lower mis-segregation of CEN V-GFP (Figure 1Aii). The CEN V-GFP spots were also incorrectly partitioned in cdc15-2 cells when released from Noc to the semirestrictive temperature of 32°C instead of 37°C (see below), with the wild-type showing a lower percentage of mis-segregation than at 37°C (see below). We furthered our studies on the cdc15-2 at 32°C instead of 37°C as the cells showed better viability at this temperature on the heated-stage during time-lapse microscopy. Consistent with the chromosome mis-segregation defect, we found that cdc15-2 cells lost viability upon a short exposure to Noc followed by recovery at the semirestrictive temperature (Figure 1B, black line).
Figure 1.
cdc15-2 cells exhibit CEN V-GFP segregation defects when exposed to nocodazole (Noc). (Ai) Logarithmic cultures of two CEN V-GFP SPC29-RFP MYO1-Redstar2 and cdc15-2 CEN V-GFP SPC29-RFP MYO1-Redstar2 cells at 24°C were shifted directly to 37°C for 5 h (left) or exposed to Noc (15 μg/ml) for 5 h before releasing into 37°C for 60 min. (Aii) Bar chart representing the percentage of cells with mis-segregated CEN V-GFP spots. (B) Logarithmic cultures of wild-type and cdc15-2 cells were diluted to OD600 = 0.1 and exposed to 15 μg/ml Noc for varying lengths of time. Viability was taken as a percentage of the number of colonies formed after exposure to Noc to the number of colonies formed in the untreated control (0 h). (C) MAD2-GFP NDC80-Redstar2 and cdc15-2 MAD2-GFP NDC80-Redstar2 cells were examined at the Noc-arrested stage (i) for the percentage of cells showing Mad2p-GFP signals (ii).
To understand basis for the CEN V-GFP mis-segregation defect in cdc15-2 cells, we first examined if there were differences in the behavior of the kinetochores during a Noc arrest between CDC15 and cdc15-2 cells. As controls, CDC15 and cdc15-2 cells arrested at metaphase in a cdc20-depleted state with intact microtubules were also observed. CDC15 and cdc15-2 cells treated with Noc showed a subpopulation of kinetochores that had dissociated from the main cluster of kinetochores (Figure S1A, ib and iib, bottom panels). The dissociation was likely due to the loss of microtubules, as cdc20-depeleted cells arrested at metaphase with intact microtubules consistently had kinetochores that mostly remained associated among themselves (Figure S1A, ia and iia).
Moreover, a subgroup of kinetochores in CDC15 and cdc15-2 cells treated with Noc were found to have drifted away from the SPBs (Figure S1, B and C, ib and iib Noc, bottom, and magnified regions with white arrows). This was unlike the kinetochores in a cdc20-depleted state where the kinetochores were held between the SPBs (Figure S1B, i and ii, and Cia and Ciia). Consistent with a previous report (Gillett et al., 2004), our data indicated that a subpopulation of kinetochores could have “drifted” from the main cluster of kinetochores in the absence of microtubules. In support of this, time-lapse experiments of cells maintained in Noc revealed that the Spc29p-RFP and CEN V-GFP underwent dynamic movements (Figure S2Aii), albeit in close association with each other in most cases (Figure S2Ai). However, at times, the dynamic movement of the SPBs and CEN V-GFP in the cells maintained in Noc (Figure S2Bii) led to the kinetochores separating from the SPBs (Figure S2Bi, 75–79 min). Nonetheless, as the extent of kinetochore drifting away from the SPBs were similar between CDC15 and cdc15-2 cells (Figure S1, percentage counts), the chromosome mis-segregation in the cdc15-2 was not likely due to the loss association of a population of chromosomes with the main cluster of chromosomes or SPBs per se.
It was previously shown in budding yeast that the spindle assembly checkpoint component Mad2p was localized to unattached kinetochores in Noc-treated cells (Gillett et al., 2004). We next examined Noc-arrested wild-type and cdc15-2 cells for the localization of the checkpoint component Mad2p-gFP and Ndc80p-Redstar2. Ndc80p is a component of the kinetochores (Wigge and Kilmartin, 2001) that allowed us to visualize all the kinetochores as an indication of the chromosomes' location. We observed that at least one Mad2p-GFP spot in each of the wild-type or cdc15-2 cells, usually at a distance away from Ndc80p-Redstar2 signals (Figure 1Ci). Our data were consistent with a previous report showing that Mad2p-CFP signals in Noc-treated cells were localized at a distance away from Ndc80p-GFP, possibly on kinetochores detached from microtubules (Gillett et al., 2004 and Figure S1). We found ∼50.67% of wild-type (n = 227) and 59.75% of cdc15-2 cells (n = 159) with Mad2p-GFP signals in the cells exposed to Noc (Figure 1Cii). The slightly lower percentage of wild-type cells with Mad2p-GFP signals was possibly due to the weaker Mad2p-GFP signals that we observed in the wild-type cells.
When time-lapse imaging was performed to examine the dynamics of Mad2p-GFP in cells released from Noc treatment, the Mad2p-GFP signals disappeared in the wild-type cells upon SPB separation (Figure S3i) in 93.2% of the cells (Figure S3iii). In contrast, cdc15-2 cells showed persistent Mad2p-GFP signals in cells where Spc29p-RFP had already separated (Figure S3ii) in 30.5% of the cells (Figure S3iii). The time-lapse images (Figure S3ii) further revealed that the Mad2p-GFP signals that were found at a distance away from the SPBs (Figure 1Ci) were not likely to be signals on the SPBs (Gillett et al., 2004) as the dynamics of the separation should have followed that of the Ndc80p-Redstar2 if indeed they were on SPBs. Taken together, the data indicated that the chromosome segregation defect in the cdc15-2 mutant was not likely due to an overt failure to activate the spindle assembly checkpoint upon Noc exposure.
cdc15-2 Cells Exhibit Lagging Chromosomes during Metaphase to Anaphase Transition after Noc Treatment
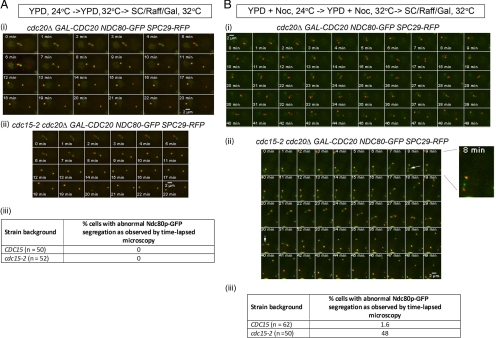
We next observed the dynamics of chromosome segregation in the mutant during metaphase-to-anaphase transition. To this end, we constructed cdc20Δ GAL-CDC20 and cdc15-2 cdc20Δ GAL-CDC20 strains carrying NDC80-GFP SPC29-RFP. The cdc20Δ GAL-CDC20 background in the CDC15 and cdc15-2 cells enabled us to compare kinetochore dynamics in cells released from Noc treatment with cells released from metaphase with intact microtubules when arrested by cdc20 depletion. Spc29p, a component of the SPBs (Elliott et al., 1999), allowed for the observation of the relative movements of the kinetochores.
During a Noc arrest, CDC15 and cdc15-2 cells exhibited two main foci of Ndc80p-GFP signals very close to the Spc29p-RFP spots (Figure 2Bi, 0 min, and Bii, 0 min; Gillett et al., 2004; Tytell and Sorger, 2006). On release, Ndc80p-GFP foci in the wild-type cells partitioned into two discrete spots where Spc29p-RFP spots have separated a distance of >2 μm (Figure 2Bi, 43–49 min). In the cdc15-2 cells, however, we consistently (Figure 2Biii) saw lagging Ndc80p-GFP signals in-between the spindle pole bodies in cells where the Spc29p-RFP signals were >2 μm apart (Figure 2Bii, 8 min, arrow).
Figure 2.
cdc15-2 cells show lagging Ndc80p signals during chromosome segregation after release from Noc. (A) cdc20Δ GAL-CDC20 NDC80-GFP SPC29-RFP (i) and cdc15-2 cdc20Δ GAL-CDC20 NDC80-GFP SPC29-RFP (ii) cells were arrested in YPD for 2.5 h at 24°C and YPD for 2.5 h at 32°C before being transferred into SC/Raff/Gal and imaged on the heated stage. The percentages of cells showing abnormal Ndc80p-GFP segregation are shown in panel iii. (B) The CDC15 (i) and cdc15-2 (ii) strains were treated similarly as in A except that Noc was added during the arrest stage in YPD. The percentages of cells showing abnormal Ndc80p-GFP segregation are shown in panel iii.
As our control, cells were arrested in metaphase with intact microtubules by CDC20 depletion. In metaphase, both wild-type and mutant cells showed the typical bilobed Ndc80p-GFP signals between the Spc29p-RFP spots (He et al., 2000; Figure 2A, i and ii, 0 min). On release from the cdc20-arrest into YP/Raff/Gal media at 32°C, wild-type and cdc15-2 cells recovered from the metaphase arrest (Figure 2Aiii), with the Ndc80p-GFP signals collapsing into discrete spots close to Spc29p-RFP, which then separated into the opposite poles of the cells (Figure 2A, i and ii, 23 min). cdc15-2 cells therefore segregated Ndc80p-GFP normally when released from a metaphase arrest with intact microtubules (i.e., cdc20-arrest) but not upon release from the Noc treatment. This supported the idea that chromosome segregation was abnormal during recovery from the spindle checkpoint activation in the mutant.
CEN V-GFP Mis-Segregation in cdc15-2 Cells Correlates with Rapid Separation of SPC29-RFP in the Presence of Unattached Kinetochores
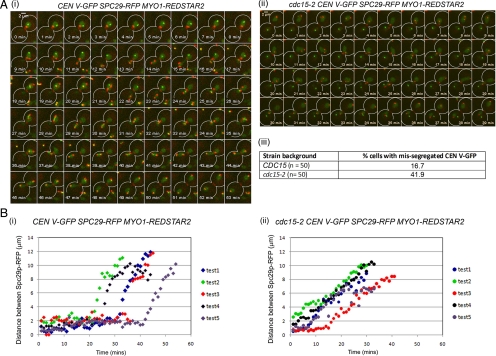
We next examined the dynamics of a single kinetochore CEN V-GFP and Spc29p-RFP separation during recovery from the spindle checkpoint activation to determine the nature of the chromosome segregation defects in the mutant. CDC15 tetR-GFP CEN5∷tetO2X112 SPC29-RFP MYO1-Redstar2 cells released from Noc into 32°C were able to successfully capture the CEN V-GFP between the Spc29p-RFP (Figure 3Ai, 27 min) and separate the pair of CEN V-GFP dots into opposite poles (Figure 3Ai, 45 min), with only 16.7% mis-segregating CEN V-GFP (Figure 3Aiii). In the cdc15-2 background, however, the Spc29p-RFP separated before the CEN V-GFP were properly captured between the SPBs (Figure 3Aii, 31 min). The mis-segregation of CEN V-GFP during release from a Noc treatment (Figure 3Aii, 39min) occurred in ∼41.9% of the cells (Figure 3Aiii).
Figure 3.
Altered dynamics of Spc29p-RFP separation in cdc15-2 cells released from Noc. (A) Wild-type tetR-GFP CEN5∷tetO2X112 SPC29-RFP MYO1-Redstar2 (i) and cdc15-2 tetR-GFP CEN5∷tetO2X112 SPC29-RFP MYO1-Redstar2 (ii) cells were arrested in Noc and released into 32°C, and time-lapse imaging carried out to examine the cells. Counts of cells with mis-segregated CEN V-GFP spots represented as a percentage of total cells examined in time-lapse microscopy are shown in panel iii. (B) The dynamics of Spc29p-RFP separation in five typical wild-type (Ai) and five typical cdc15-2 (Aii) cells were determined by measuring the distance between Spc29p-RFP and plotted over time.
We noted that the movement of the CEN V-GFP and Spc29p-RFP spots was rather dynamic during Noc recovery in wild-type and cdc15-2 cells (Movies 1 and 2 from Figure 3A, i and ii, respectively), reminiscent of the “oscillatory” movements of the nucleus previously described (Yeh et al., 1995). The Spc29p-RPF appeared to transit between compartments (Movies 1 and 2). We speculate that this was due to the cells trying to establish proper orientation and association of the cytoplasmic microtubules with the cortex upon recovery from Noc arrest. Indeed, the oscillatory movement in both CDC15 and cdc15-2 cells was absent or less dramatic in cells released from a Cdc20p depletion (Figure S4), presumably as the spindles were intact in the absence of Cdc20p function. On restoration of Cdc20p expression, Pds1p destruction ensued, and an extension of spindles already attached to the kinetochores occurred. As such, there was no requirement to neither orientate nor capture kinetochores and hence an absence of oscillations when recovering from a Cdc20p-depleted state. We wondered if the oscillations during the recovery phase resulted in the failure of the mutant cells to capture the CEN V-GFP that had drifted away (Figure S1), thereby causing the mutant cells to mis-segregate chromosomes. Our observations indicated that there were no direct correlations between the transition of Spc29p-RFP across the neck and the mis-segregation of chromosomes in the cdc15-2 mutant (data not shown).
We then examined the dynamics of spindle elongation during recovery from Noc by measuring the separation of the Spc29p-RFP spots, which we used as an indicator for spindle elongation. In five typical wild-type cells, the Spc29p-RFP spots maintained at <4 μm apart for a duration that ranged from 20 to 40 min upon release from Noc (Figure 3Bi), before separating the Spc29p-RFP and CEN V-GFP. In contrast, five cdc15-2 cells examined showed Spc29p-RFP spots separating relatively quickly, on average within 25 min after release from Noc without the proper capture and/or biorientation of the CEN V-GFP spots (Figure 3Bii). Our data showed a correlation between uninhibited spindle elongation in the presence of unattached kinetochores and aberrant CEN V-GFP segregation during inactivation of the spindle checkpoint in the cdc15-2 cells.
cdc15-2 Cells Achieve Chromosome Biorientation during Recovery from Noc Release if Spindle Elongation is Restrained
Next we asked if cdc15-2 cells could capture and biorientate the chromosomes correctly under conditions where the spindle length was constrained. We made use of CDC15 and cdc15-2 strains carrying cdc20Δ GAL-CDC20 tetR-GFP CEN5∷tetO2X112 SPC29-RFP, which allowed us to restrain spindle elongation by maintaining cells in the cdc20-arrested state after release from Noc. If the wild-type and cdc15-2 cells were able to biorientate sister chromatids when the mitotic spindles were short in the cdc20-arrest subsequent to Noc washout, we would be able to observe the CEN V-GFP spots alternating between two separated spots and one collapsed spot (centromere breathing; He et al., 2000).
As a control, cdc20Δ GAL-CDC20 and cdc15-2 cdc20Δ GAL-CDC20 cells were observed at the cdc20 arrest with intact spindles. CDC15 (Figure S5A) and cdc15-2 cells (Figure S5B) exhibited similar abilities (Figure S5C) to biorientate chromosomes, supporting the idea that chromosome mis-segregation occurred in the cdc15-2 mutant only upon recovery from Noc exposure. Interestingly, the cells with the cdc15-2 arrested at the Cdc20p depletion appeared to have Spc29p-RFP at a further distant apart from each other (Figure S5E) than in CDC15 cells (Figure S5D). When we measured the distance between the SPBs, we found on average a longer distance between the Spc29p-RFP in the cdc15-2 cells (Figure S5F). We examined the Pds1p levels in a Cdc20 arrest in parallel experiments using CDC15 and cdc15-2 cdc20Δ GAL-CDC20 CEN5∷tetO2X112 SPC29-RFP PDS1-HA cells. The longer spindle length in the cdc15-2 background correlated with the slightly lower Pds1p-HA levels in the mutant (Figure S5G), indicating that cdc15-2 may have a defect in Pds1p accumulation (see below).
CDC15 and cdc15-2 cdc20Δ GAL-CDC20 tetR-GFP CEN5∷ tetO2X112 SPC29-RFP strains were next arrested in YPD containing Noc, following which the Noc was washed out and cells were resuspended in YPD to allow for reformation of spindles but in the absence of Cdc20p. This will restrain SPB separation during Noc recovery. If the cdc15-2 cells from a Noc treatment released into the cdc20 arrest were able to capture and biorientate sister chromatids when the mitotic spindles were short, we would be able to observe breathing of the CEN V-GFP spots between separated Spc29p-RFP.
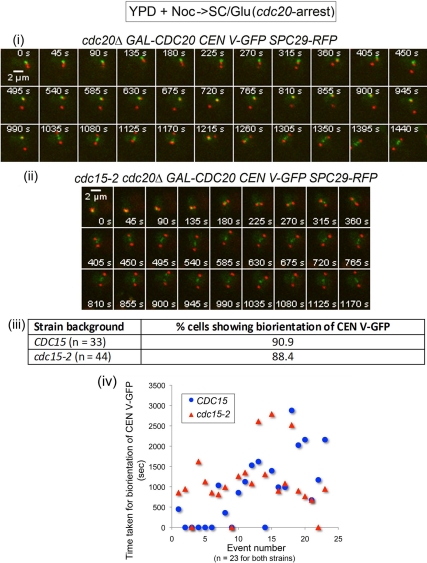
In the control cdc20Δ GAL-CDC20 cells (n = 33), the CEN V-GFP spots were captured subsequently by one of the SPB and eventually biorientated after association with the first SPB as evident from the separation of the CEN V-GFP spots (Figure 4i, 1350–1440s). This is consistent with a report showing that budding yeast chromosomes tend to biorientate chromosomes once a kinetochore associates with a SPB (Indjeian and Murray, 2007). cdc15-2 cdc20Δ GAL-CDC20 cells (n = 44) released from Noc into the cdc20 arrest showed biorientation (Figure 4ii, 450-1170 s) in close to 90% of the cells (Figure 4iii). The association between the kinetochores and microtubules was stable in the cdc15-2 mutant as we hardly observe loss of biorientation subsequent to the initial capture (data not shown).
Figure 4.
Restraining spindle elongation in cdc15-2 cdc20Δ GAL-CDC20 tetR-GFP CEN5∷ tetO2X112 SPC29-RFP resulted in proper biorientation of CEN V-GFP. Wild-type (i) and cdc15-2 (ii) cells were arrested in YPD + Noc and released into SC/Glu to maintain cells in metaphase as they recover from Noc during time-lapse imaging. (iii) Percentage of cells showing proper biorientation. (iv) Plots show the time taken to biorientate CEN V-GFP in CDC15 and cdc15-2 cells. Time needed for biorientation was taken as the time between Spc29p-RPF separation, and CEN V-GFP stretched across the separated Spc29p-RFP.
We also examined closer the kinetics of biorientation in these cells arrested at the Cdc20p-depleted stage. We defined the time taken to biorientate CEN V-GFP as the time after the SPBs have visibly separated to the time the CEN V-GFP was first seen to be stretched across the Spc29p-RFP. For instance, in Figure 4i, the time point at which the Spc29p-RFP became visibly separated was at 315 s, whereas the time point at which the CEN V-GFP was seen stretched across the SPBs was at 1035 s. The time taken for biorientation in this cell was therefore 720 s. There appeared to be no significant difference in the time needed for cells to biorientate CEN V-GFP in the wild-type and mutant cells (Figure 4iv, n = 23 for both strains). Our data indicated that the chromosome mis-segregation in the mutant cells was not due to defective kinetochore attachment per se, but that untimely spindle elongation (Figure 3) could have led to the chromosome segregation defect.
Chromosome Segregation Defect in cdc15-2 Is Rescued by a Deletion in CIN8
It was previously shown that the passenger protein Ipl1p plays a role in the biorientation of sister chromatids (Biggins and Murray, 2001; Tanaka et al., 2002). Ipl1p is also needed for detecting the loss of cohesion between sister chromatids and activating the spindle assembly checkpoint (Biggins and Murray, 2001). Cdc15p was previously found to be required for the proper localization of Ipl1p and Sli15p (Stoepel et al., 2005). We sought to determine if Ipl1p localization were altered in the cdc15-2 mutant during Noc recovery at 32°C that could have contributed to the mis-segregation of CEN V-GFP upon Noc release. Sli15p, a related kinetochore passenger protein that is a known substrate of Ipl1p (Kang et al., 2001; Cheeseman et al., 2002), was also examined.
Ipl1p-GFP was localized to the spindles in wild-type (n = 20; Figure S6Ai) and the cdc15-2 cells (n = 22; Figure S6Aii). However, the Ipl1p-GFP signals decorating the spindles in mutant cells indicated that the spindles were curved (Figure S6Aii, 66 min, white arrow) in 15 of 22 cells compared with two in 20 wild-type cells. The bent spindles resulted in the asymmetric positioning of the midzone (as indicated by the region on the spindle where Ipl1p-GFP signal was more intense; Buvelot et al., 2003) relative to the constriction plane (Figure S6Aii, 66–72 min, white asterisks). On spindle breakdown (Figure S6Aii, 72 min, white arrow), both the plus ends of the microtubules, as marked by the intense Ipl1-GFP staining, were found in the same compartment (Figure S6Aii, 72 min, white arrow).
In CDC15 (n = 21), Sil15p-GFP was localized to the spindles (Figure S6Bi). At the point of Myo1p-Redstar2 ring constriction, Sli15p-GFP started to dissociate from the midzone and spindles (Figure S6Bi, 44 min, white asterisk). In cdc15-2 cells (n = 25), Sli15p-GFP was also localized onto the spindles (Figure S6Bii). From the Sli15p-GFP signals, it could also be seen that the spindles were bent (Figure S6Bii, 40–42 min, white arrow) in 17 of 25 cdc15-2 cells compared with 3 of 21 in wild-type cells. The mis-positioned midzone was also obvious in the mutant cells (Figure S6Bii, 48 min, white arrow). However, the localization of Sli15p at the midzone in the mutant appeared somewhat defective (Figure S6Bii).
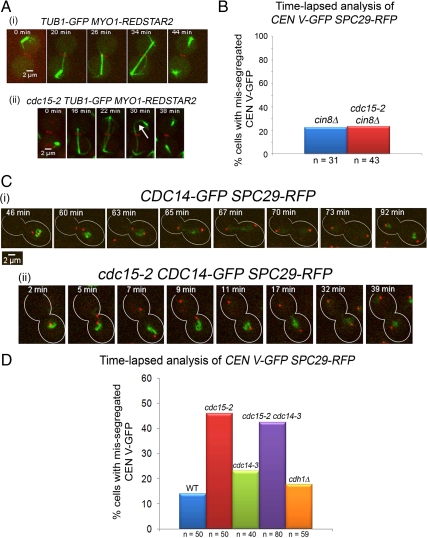
The premature spindle elongation (Figure 3Bii) together with the Ipl1p and Sli15p signals decorating the spindles in the mutant cells indicated that spindle dynamics was abnormal in the cdc15-2 cells. Indeed, Tub1p-GFP in the mutant cells released from Noc arrested consistently showed spindles were abnormally long, leading to a curved appearance (Figure 5Aii, 30 min, white arrow). This was unlike that in the wild-type cells released from Noc where the spindles were typically straight (Figure 5Ai, 34 min).
Figure 5.
(A) Abnormal microtubule dynamics in cdc15-2 cells released from Noc. Wild-type (i) and mutant (ii) cells carrying TUB1-GFP MYO1-REDSTAR2 were arrested in Noc, released into 32°C, and examined using time-lapse microscopy. Selected time points are shown. (B) CIN8 deletion rescued CEN V-GFP mis-segregation in cdc15-2 cells. cin8Δ tetR-GFP CEN5∷tetO2X112 SPC29-RFP and cdc15-2 cin8Δ tetR-GFP CEN5∷tetO2X112 SPC29-RFP cells were treated as described in Figure 4 and the number of cells with mis-segregated CEN V-GFP are represented as a percentage of the total number of cells counted. (C) Cdc14p-GFP is not prematurely released in cdc15-2 cells. CDC15 CDC14-GFP SPC29-RFP (i) and cdc15-2 CDC14-GFP SPC29-RFP (ii) cells were treated as described in Figure 4. (D) Mis-segregation of CEN V-GFP in cdc15-2 cells not rescued by cdc14-3. Strains with various genotypes as indicated were treated as described in Figure 3, and the number of cells with mis-segregated CEN V-GFP are represented as a percentage of the total number of cells counted.
To determine if the spindle elongation defect led to the chromosome mis-segregation in the cdc15-2 cells, we tested if a deletion of the genes encoding for motor proteins needed for spindle elongation would rescue the defect. Cin8p is a microtubule-associated kinesin-5 protein and has been observed on the SPBs, kinetochores, spindles, and midzone (Schuyler et al., 2003; Tytell and Sorger, 2006; Gardner et al., 2008). Cin8p has been shown to be required for spindle elongation (Saunders et al., 1997), specifically for the rapid phase of mitotic spindle elongation (Straight et al., 1998). The ectopic expression of Cin8p (Krishnan et al., 2004; Liu et al., 2008) has been shown to promote spindle elongation.
cin8Δ tetR-GFP CEN5∷tetO2X112 SPC29-RFP and cin8Δ cdc15-2 tetR-GFP CEN5∷tetO2X112 SPC29-RFP cells were arrested in Noc and released from Noc as described for the experiments in Figure 3. Interestingly, cin8Δ tetR-GFP CEN5∷tetO2XS112 SPC29-RFP and cin8Δ cdc15-2 tetR-GFP CEN5∷tetO2X112 SPC29-RFP cells showed similar levels of CEN V-GFP mis-segregation (Figure 5B), indicating that a cin8Δ rescued the CEN V-GFP mis-segregation defect in the cdc15-2 cells. Our data supported the idea that the unrestrained spindle elongation during recovery from Noc led to the inability of the cdc15-2 mutant to properly capture chromosomes, perhaps due to untimely localization of Cin8p.
Mis-Segregation of CEN V-GFP in cdc15-2 Cells not Likely due to Premature Activity of Cdc14p or Compromised Function of Cdh1p
It was shown that Cdc14p stabilizes microtubule dynamics (Higuchi and Uhlmann, 2005), thereby promoting anaphase (Higuchi and Uhlmann, 2005). Furthermore, it was recently shown that the spindle assembly checkpoint acts on Cdc14p to prevent premature separation of the SPBs (Chiroli et al., 2009). We asked if the defect in cdc15-2 cells were related to a loss of proper regulation of Cdc14p resulting in the abnormal spindle elongation during Noc release and whether Cdc14p-GFP was released prematurely from the nucleolus (Geymonat et al., 2002) in cdc15-2 cells in the presence of unattached kinetochores.
CDC14-GFP SPC29-RFP cells released from Noc separated Spc29p-RFP only after Cdc14p-GFP was released from the nucleolus as seen from the reduction in the GFP signals (Figure 5Ci, 65 min, n = 10). In the cdc15-2 background, we observed cells (n = 10) in which the Spc29p-RFP separated quickly that Cdc14p-GFP was observed largely to be still in the nucleolus (Figure 5Cii, 7–17 min). Cdc14p-GFP was released from the nucleolus only at 32 min (Figure 5Cii), when the SPBs had long separated.
We also tested if inactivating Cdc14p function by introducing a cdc14-3 mutation into cdc15-2 CEN V-GFP SPC29-RFP cells could rescue the chromosome mis-segregation defect. Time-lapse analysis showed that the CEN V-GFP mis-segregation in cdc15-2 cdc14-3 (42%) was not significantly less than in the cdc15-2 cells (46%; Figure 5D), indicating that the mis-segregation of CEN V-GFP in cdc15-2 was probably not because of premature activation of Cdc14p function, leading to early SPB separation relative to chromosome segregation (Chiroli et al., 2009). These observations supported the time-lapse data (Figure 5C) showing that the defect in cdc15-2 was not likely due to premature activation of Cdc14p function leading to spindle elongation.
Cdc15p is a component of the MEN that promotes the activation of Cdh1p (see above). Although at 32°C cdc15-2 cells exit mitosis (Hwa Lim et al., 2003), the mitotic exit is slightly sluggish (data not shown), indicating that Cdh1p may not be fully active. Interestingly, Cdh1p has previously been implicated in maintaining genomic stability (Ross and Cohen-Fix, 2003). Also, Cin8p, a substrate of the APCCdh1p (Hildebrandt and Hoyt, 2001), could have accumulated in the cdc15-2 mutant due to improper regulation of the Cdh1p function.
Examination of CEN V-GFP segregation in cdh1Δ cells revealed that the cdh1Δ cells did not show a higher number of mis-segregation of CEN V-GFP compared with wild-type cells (Figure 5D). As such, the defect observed in the cdc15-2 mutant could be unrelated to its mitotic exit function with respect to Cdh1p. The defective chromosome segregation in cdc15-2 cells upon Noc recovery may therefore be due to a specific role of Cdc15p earlier on in mitosis.
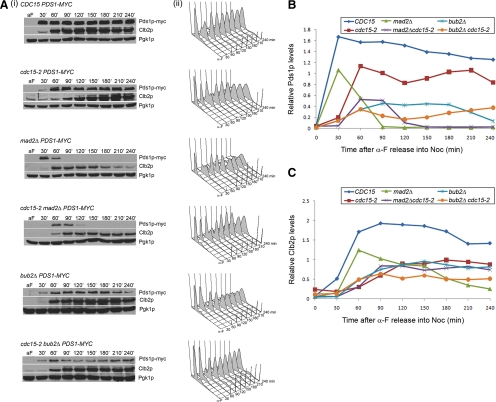
Pds1p Fails to Accumulate to the Same Extent in cdc15-2 Cells Compared with Wild-Type Cells
From our microscopic examination, we found that cdc15-2 cells appeared to arrest as dumb-bells (e.g., Figure 1C) with unseparated Spc29p-RFP (e.g., Figure S1B) when treated with Noc. However, given the chromosome mis-segregation defect, we examined more closely Pds1p-myc levels during Noc arrest. Wild-type and cdc15-2 cells accumulated Pds1p-myc and Clb2p levels when they were released from α-factor into Noc (Figure 6Ai), indicating that the cells could activate the spindle assembly checkpoint in response to unattached kinetochores unlike in mad2Δ cells (Figure 6Ai). Interestingly, the Pds1p levels in cdc15-2 (Figure 6B, red line) cells did not accumulate to the same extent as in wild-type cells (Figure 6B, blue line). The FACs profile (Figure 6Aii) of the cdc15-2 cells indicated that the mutant was progressing into mitosis slower (Figure 6, 120 min) than the wild-type cells (Figure 6Aii, 60 min).
Figure 6.
Pds1p levels do not accumulate as high in cdc15-2 cells. (A) Cells from various strains were sampled at 30 min-intervals after release from α-factor into Noc at 32°C for 4 h. Western blot analysis of Pds1p-myc, Clb2p, and loading control Pgk1p are shown in panel i. FACs profiles of the cells at each time point are shown in panel ii. (B) Densitometry readings of the Western blot showing the ratio of Pds1p levels to Pgk1p levels. (C) Densitometry readings of the Western blot showing the ratio of Clb2p levels to Pgk1p levels.
The spindle assembly checkpoint has two arms, one dependent on Mad2p (Musacchio and Salmon, 2007) and the other on Bub2p (Tan et al., 2005). The cdc15-2 mad2Δ mutant showed a lower level of Pds1p accumulation (Figure 6, Ai and B, purple line) than either cdc15-2 or mad2Δ mutant (Figure 6B, green line). Bub2p plays a role in the preventing the activation of Tem1p, the MEN component upstream of Cdc15p, in the presence of Noc (Tan et al., 2005). Pds1p level persisted longer in the bub2Δ mutant compared with the mad2Δ. This was consistent with a previous study showing that Bub2p is not essential for Pds1p stabilization during a Noc arrest (Alexandru et al., 1999). We noted that the levels of Pds1p in cdc15-2 bub2Δ double mutant accumulated slower (Figure 6B, orange line) than in the bub2Δ (Figure 6B, cyan line) single mutant. Also, the FACs profile showed that the cdc15-2 background in mad2Δ or bub2Δ slowed the progression into mitosis relative to the single mutants (Figure 6Aii).
The delay in cell cycle progression was also reflected in the lower level of Clb2p accumulation in the cdc15-2 cells compared with the wild-type cells (Figure 6, Ai and C). Likewise, Clb2p accumulation was delayed in the mad2Δ or bub2Δ with a cdc15-2 background (Figure 6, Ai and C). Our observations would suggest that the slower transition through the cell division cycle in the cdc15-2 mutant led to a deficiency in the levels of Pds1p accumulation during a Noc arrest, leading to a less than optimal Pds1p level that likely affected recovery from the spindle assembly checkpoint activation.
Increasing Pds1p Levels Can Rescue CEN V-GFP Mis-Segregation Defect in cdc15-2 Cells
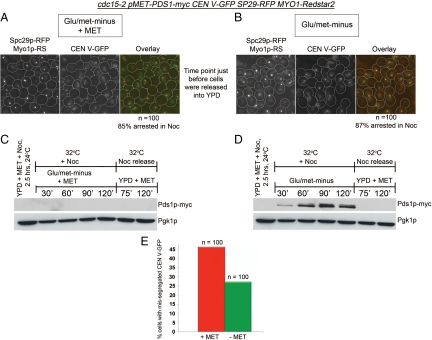
To confirm if a lowered level of Pds1p was contributing to the mis-segregation of CEN V-GFP in the cdc15-2 background, we constructed a cdc15-2 tetR-GFP CEN5∷tetO2X112 SPC29-RFP MYO1-Redstar2 strain carrying pMET-PDS1myc, which allowed us to overproduce Pds1p-myc during a Noc arrested by culturing the cells in methionine-minus medium. The strains were cultured in YPD+MET+Noc at 24°C for 2.5 h. The cells were quickly spun down, and half the culture was resuspended in Glu/−met medium+Noc+ MET as control and the other Glu/−met medium+Noc for de-repression of the MET3 promoter. After 2 h at 32°C, the cultures were spun down and released into YPD +MET at 32°C and CEN V-GFP segregation examined. The experimental regime is shown in Figure S7.
Figure 7, A and B, shows that at least 85% of the cells arrested in Noc before the cells were released into YPD. In the presence of methionine in the Met− medium, the control culture did not show Pds1p-myc expression (Figure 7C). When methionine was absent, Pds1p-myc was expressed from 30 to 120 min after de-repression (Figure 7D). On Noc release, the control cells where the MET3 promoter was repressed by the presence of methionine (+Met) showed ∼46% of CEN V-GFP mis-segregation (Figure 7E). In the culture where the MET3 promoter was de-repressed (−Met), we observed close to a 50% decrease in CEN V-GFP mis-segregation (Figure 7E). Our data implied that a failure to accumulate Pds1p levels at a critical level during Noc arrest could result in premature spindle elongation and mis-segregation of chromosomes upon Noc release.
Figure 7.
cdc15-2 tetR-GFP CEN5∷tetO2X112 SPC29-RFP MYO1-Redstar2 MET-PDS1myc cells showed reduction in CEN V-GFP mis-segregation during Noc release when Pds1p was overproduced during Noc arrest. An overnight culture of the strain was arrested in Noc in YPD at 24°C for 2.5 h. The culture was then divided into two, with one-half of the culture resuspended in Glu/met− medium containing methionine (+Met) to repress the MET promoter driving PDS1myc expression and Noc. The second half was resuspended in methionine-minus medium (−Met) to de-repress the MET promoter in the presence of Noc. After 2 h at 32°C, both cultures were released into YPD. The cells after 2 h in Glu/met− with methionine (A) and Glu/met− (B) are shown together with the percentages of Noc-arrested cells. The percentages were determined from the number of cells from a total of 100 cells with both SPBs collapsed together as a single spot. Western blot analysis of the lysates taken at the indicated time points show the level of Pds1p-myc in the presence (C) or absence (D) of methionine. (E) Graph shows percentage of telophase cells with mis-segregated CEN V-GFP in +Met or −Met medium at 75 min after release from Noc.
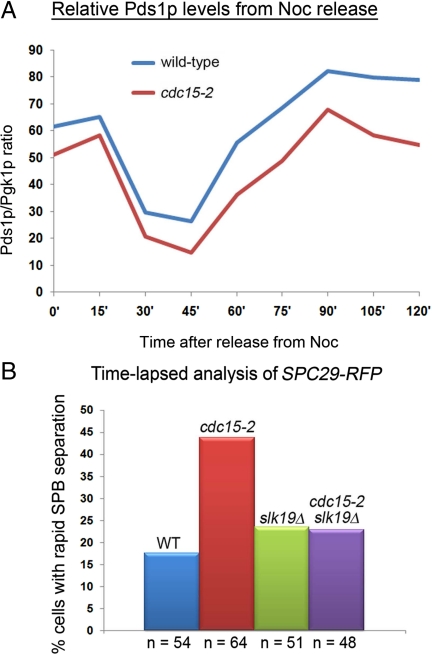
SLK19 Deletion Rescued Rapid SPB Separation in cdc15-2 Cells
The Pds1p levels while low in the cdc15-2 cells (Figure 6) was perhaps maintained at a threshold level sufficient for the arrest (Figure 1B). When we observed the levels of Pds1p-myc in wild-type and cdc15-2 cells arrested in Noc from cycling cultures, we consistently detected lower levels of Pds1p-myc (Figure 8A), although both strains were arrested for 5 h. Interestingly, the dynamics of the Pds1p degradation as judged from the slope of the graph during Noc release appeared similar for wild-type and cdc15-2 cells, but the lower starting levels of Pds1p levels meant that Pds1p levels decreased faster in the cells (Figure 8A).
Figure 8.
(A) Wild-type and cdc15-2 cells were released from Noc-arrest into YPD at 32°C. Graph shows the levels of Pds1p relative to Pgk1p at each time point. (B) Deletion of SLK19 rescued rapid SPB separation phenotype in cdc15-2 cells. Cells from various strains as indicated were treated as in Figure 3. The plot shows the percentage of cells showing fast SPB separation as examined by time-lapse imaging.
The effect of the compromised Pds1p level becomes apparent upon Noc release as evident from the mis-segregation of CEN V-GFP (Figure 4Aii). Separase (Esp1p), which is normally inhibited by Pds1p (Ciosk et al., 1998), could be relatively uninhibited due to the lower Pds1p levels. Separase is involved in spindle elongation (Sullivan and Uhlmann, 2003; Baskerville et al., 2008). As a result of the less than optimal inhibition by Pds1p, the separase could promote untimely spindle elongation in the mutant cells, leading to the uncoordinated spindle elongation and chromosome segregation. A previous study also showed that spindle stabilization and elongation requires both activation of the APCCdc20p and chromosome segregation, but the activity of separase was dispensable (Severin et al., 2001). However, it was shown in other studies that separase activity is needed (Baskerville et al., 2008).
Separase functions together with Slk19p (Sullivan et al., 2001), a kinetochore and spindle midzone protein (Zeng et al., 1999), during anaphase when spindle elongation occurs (Khmelinskii et al., 2007). We examined SPB separation in wild-type, cdc15-2, slk19Δ, or cdc15-2 slk19Δ cells harboring SPC29-RFP to ascertain if an SLK19 deletion could rescue the premature spindle elongation defect. As can be seen, the percentage of cells with rapid SPB separation occurring within 25 min after Noc release (Figure 4Bii) in the cdc15-2 slk19Δ cells was similar to the slk19Δ cells (Figure 8B), suggesting that the rapid SPB separation in the cdc15-2 mutant could be due to aberrant regulation of Slk19p, leading to the premature stabilization of the midzone or recruitment of motor proteins such as Cin8p, to the spindles (Khmelinskii et al., 2007), thereby promoting spindle elongation before proper attachment of kinetochores had occurred. Consistent with this, a cin8Δ rescued the CEN V-GFP mis-segregation in the cdc15-2 mutant (Figure 5B).
Cin8p-GFP Localizes to Interpolar Microtubules in cdc15-2 Cells Showing Rapid Spc29p Separation
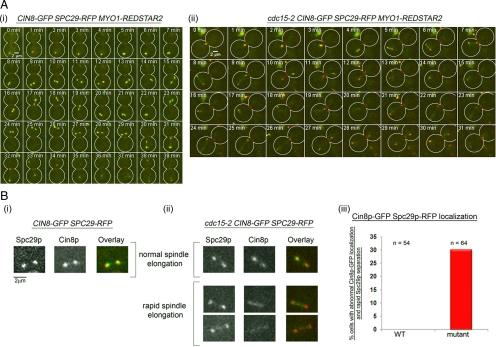
Prompted by our observations that either a deletion in CIN8 or SLK19 led to a rescue in the mis-segregation of CEN V-GFP (see above), we performed time-lapse imaging in wild-type CIN8-GFP MYO1-Redstar2 and cdc15-2 CIN8-GFP MYO1-Redstar2 cells to directly examine if there were differences in the dynamics of Cin8p behavior in the mutant cells that could account for the rapid spindle elongation and CEN V-GFP mis-segregation (Figure 3).
We observed that Cin8p-GFP localized in a similar pattern in both wild-type and cdc15-2 cells upon Noc release (Figure 9Ai). Typically, the Cin8p-GFP signals were close to and overlapping with that of the Spc29p-RFP spots in cells with short spindles (Figure 9, Ai, 8–29 min, and Bi). In cells elongating their spindles, Cin8p-GFP could be observed on the interpolar spindles (Figure 9Ai, 30 min). These observations were consistent with previous observations (Khmelinskii et al., 2007, 2009; Gardner et al., 2008). However, we noted that in a subpopulation of mutant cells that the Cin8p-GFP signals appeared at the interpolar region between the Spc29p-RFP fairly quickly after Noc release (Figure 9Aii, 5–6 min). Closer examination revealed that the mutant cells that showed such interpolar localization of Cin8p-GFP at the early time point after Noc release had separated Spc29p-RFP rapidly (Figure 9B, ii, bottom panel, and iii).
Figure 9.
Cin8p-GFP localizes prematurely in cdc15-2 cells. (A) CIN8-GFP SPC29-RFP (i) and cdc15-2 CIN8-GFP SPC29-RFP (ii) cells were released from Noc arrest to YPD at 32°C as described in Figure 3 and time-lapse images taken. (B) Magnified images of Cin8p- Spc29p-RFP in wild-type (i) and mutant (ii, top panel) cells with normal timing of Spc29p-RFP separation. Premature localization of Cin8p-GFP to interpolar microtubules was observed in cdc15-2 cells (ii, bottom panel). (iii) Graph showing percentages of wild-type and mutant cells with premature Cin8p-GFP localization to interpolar microtubules in cells with rapid Spc29p-RFP.
Interestingly, it was recently found that the recruitment of Cin8p in a separase-dependent manner to the interpolar region is needed to drive spindle elongation (Khmelinskii et al., 2009). Our data showing rapid separation of Spc29p-RFP in cdc15-2 cells with CEN V-GFP mis-segregation (Figure 4) was consistent with a mis-regulation of Cin8p localization upon Noc release due to lower Pds1p levels in the mutant.
DISCUSSION
One of the key events during mitosis is the biorientation of sister chromatids before their proper segregation. During the early phase of a cell division cycle, the kinetochores are near to the SPBs by their association with the spindles (Winey and O'Toole, 2001), such that during a normal metaphase or in the event that Cdc20p were depleted, the kinetochores would remain attached to the intact microtubules. This tight association of the kinetochores with the SPBs (Figure S1) is reflected in the “centromere breathing” that is observed in cells held at a cdc20-arrest (Figure S5; He et al., 2000). Also, because of the geometric constraints of the kinetochores on sister chromatids, the kinetochores on a pair of sister chromatids have a propensity to biorientate as long as one of them is attached to microtubules (Indjeian and Murray, 2007) before metaphase. In a normal mitosis, therefore, biorientation of sister chromatids would have occurred before complete elongation of the spindles.
In cells exposed to Noc, however, the loss of microtubules can cause kinetochores to drift away from the SPBs (Figure S1B and S2B). Such kinetochores are captured in the wild-type cells before the separation of the SPBs (Figure 3Ai). In cdc15-2 cells, however, the kinetochores that have drifted away from the SPBs failed to be captured and biorientated properly when Noc was removed (Figure 3Bii), resulting in mis-segregation of sister chromatids. Intriguingly, cdc15-2 cells in Noc arrested as large-budded cells and showed Mad2p localization (Figure 1B), indicating that the spindle assembly checkpoint was functional. Indeed, time-lapse imaging of Mad2p-GFP revealed that Mad2p-GFP persisted in cdc15-2 cells at a distance away from the main cluster of kinetochores as marked by Ndc80p-Redstar2 (Figure S3ii). However, the Mad2p-GFP signals on kinetochores distant from the SPBs were not properly captured in the mutant while spindle elongation occurred.
Interestingly, cdc15-2 cells showed proper kinetochore capture (e.g., Figure S1Aiia) and chromosome segregation when the cells had no prior exposure to Noc (e.g., Figure 1Ai). This supported the notion that the chromosome mis-segregation defect in the mutant was related to premature spindle elongation (Figure 3Bii) only in cells where kinetochores have lost association with the microtubules during a Noc exposure. Indeed, restraining spindle elongation during recovery from Noc arrest (Figure 4) or abolishing the function of Cin8p in cdc15-2 cells (Figure 6B) rescued the CEN V-GFP mis-segregation, which indicated that a mis-regulation of the motor proteins might have contributed to the CEN V-GFP mis-segregation. This also implied that the mis-segregation of CEN V-GFP in cdc15-2 cells was unlikely to be related to kinetochore defects.
What then could have resulted in a loss of proper control of spindle elongation and sister chromatid segregation during recovery from spindle assembly checkpoint activation in cdc15-2 cells, given that the spindle assembly checkpoint appeared to be functional (Figure 1C and Figure S3)? We observed that Pds1p levels were not as highly accumulated in the mutant compared with the wild-type cells, as the mutant was delayed in cell cycle progression (Figure 6Ai). Also, a Cdc20 depletion in cdc15-2 cells resulted in lower Pds1p-HA levels (Figure S5G). This might seem inconsistent with the role of APCCdc20 in the degradation of Pds1p (Visintin et al., 1997). However, it is conceivable Pds1p was defective in accumulating before complete depletion of Cdc20p due to the slow transition through mitosis (Figure 6Aii). The possibility that the mis-segregation of CEN V-GFP in the cdc15-2 cells was related to the Pds1p levels was in agreement with the finding that the forced expression of Pds1p during Noc arrest in the cdc15-2 cells reduced the mis-segregation of CEN V-GFP (Figure 7E).
As a consequence of the suboptimal accumulation of Pds1p, it was probable that the separase normally held in check by Pds1p could be relatively uninhibited. The separase could therefore promote spindle elongation faster in the mutant than wild-type cells upon release from Noc due to the faster decline in Pds1p levels (Figure 8A). This could have resulted in the untimely spindle elongation in the absence of proper chromosome capture (Figure 3Aii). Direct examination of Cin8p-GFP in cdc15-2 cells revealed a good correlation between the cells showing rapid Spc29p-RFP separation and localization of Cin8p-GFP to the interpolar microtubules fairly quickly after recovery from Noc (Figure 9Bii). Recent studies (Fridman et al., 2009; Khmelinskii et al., 2009) back the idea that Cin8p localization at the interpolar region plays a role in the elongation of spindles, thereby pushing apart the SPBs during anaphase. Our observations would suggest that the premature localization of Cin8p onto short mitotic spindles (Figure 9Bii) resulted in rapid spindle elongation without the complete capture of kinetochores in the mutant (Figure 3, A and Bii).
It was previously shown that the anaphase B spindles undergo a fast and slow phase of extension, with the fast phase dependent on Cin8p (Straight et al., 1998). Time-lapsed imaging of the SPB separation in the cdc15-2 cells during Noc recovery indicated that the fast phase of spindle elongation has occurred (Figure 3Bii) in spite of the presence of unattached kinetochores (Figure 3Aii). A recent study showed that the proper localization of Cin8p depends on midzone proteins such as Esp1p and Slk19p during anaphase (Khmelinskii et al., 2007). Interestingly, abolishing the midzone protein Slk19p reduced the percentage of cdc15-2 cells showing rapid SPB separation (Figure 8B), whereas deletion of CIN8 led to a rescue in CEN V-GFP mis-segregation (Figure 5B). These findings lend support to the idea that the localization of Slk19p and Cin8p are mis-regulated in cdc15-2 cells. The localization of both these midzone proteins requires Ase1p, which in turn needs to be dephosphorylated by Cdc14p (Khmelinskii et al., 2007). However, the inability of the cdc14-3 mutation to rescue the CEN V-GFP mis-segregation in cdc15-2 (Figure 5D) indicated that the defect was upstream of Cdc14p function. In fact, it was shown that there is a Cdc14p-independent function of Esp1p-Slk19 in spindle elongation, presumably at the initial fast phase of anaphase B (Khmelinskii et al., 2007). That the initial stage of SPB separation occurred while Cdc14p-GFP remained in the nucleolus in cdc15-2 cells (Figure 5C, 5–9 min) further indicated a loss of proper control in the timely execution of Slk19p function can result in spindle elongation in the presence of unattached kinetochores. Moreover, the observation that Ipl1p-GFP and Sli15p-GFP was somewhat defective in localizing to the spindle midzone in the mutant cells (Figure S6ii) is consistent with the finding that Cdc14p-GFP was not released prematurely from the nucleolus in cdc15-2 cells elongating the mitotic spindles (Figure 5C). Further studies on the dynamics of Ipl1p and Sli15p localization might provide some answers as to the reasons underlying the failure of the cc15-2 cells to detect the absence of proper kinetochore capture and biorientation during recovery from Noc treatment.
The transcription of PDS1 peaks at G1/S (Spellman et al., 1998), whereas the Pds1p level peaks at mitosis (Tinker-Kulberg and Morgan, 1999). Our observations indicated that a slower progression through the cell division cycle in the cdc15-2 cells led to a delayed expression of PDS1 (Figure 6). Although the cdc15-2 cells eventually reached mitosis at 120 min after α-factor release and remained in mitosis till 240 min, the level of Pds1p and Clb2p did not reach the levels as that in the wild-type cells (Figure 6, B and C). This is likely due to the inefficient transcription of PDS1 and CLB2 as well as the less than synchronous passage through the cell division cycle in the mutant cells. In addition, the cdc15-2 background in either a mad2Δ or bub2Δ led to a lower Pds1p accumulation than in either single mad2Δ or bub2Δ mutants (Figure 6). The phosphorylation of Pds1p by Cdc28p (Agarwal and Cohen-Fix, 2002; Holt et al., 2008) has been shown to regulate its stability (Holt et al., 2008). The slower rate of entry into mitosis, as reflected in the Clb2p levels in the cdc15-2 mutant (Figure 6C), could have led to lower Clb2p-Cdc28p activity that contributed to the lower levels of Pds1p accumulation and hence affected the recovery from Noc arrest, even in the presence of an intact spindle assembly checkpoint. Taken together, our observations suggest that a minimal threshold level of Pds1p is needed for cells to arrest in metaphase in the presence of Noc. However, a lower level of accumulated Pds1p at the arrest stage could lead to a faster decline in Pds1p levels (Figure 8A) during recovery from the spindle checkpoint activation, resulting in a defect in the regulation of spindle elongation and mis-segregation of sister chromatids.
Our results therefore underscore the fact that preventing chromosome segregation in the event of spindle damage in cells cannot be dependent solely on the activation of the spindle assembly checkpoint. Rather another key aspect of a cell's response toward spindle defects is a timely transition through the cell division cycle and expression of PDS1 before recovery from the checkpoint activation so as to delay spindle elongation until all kinetochores are captured. The successful inheritance of chromosomes during cell division is hence a complex process that requires the concerted functions of the cell cycle components as well the spindle assembly checkpoint pathway for proper coordination of spindle elongation with chromosome segregation.
Supplementary Material
ACKNOWLEDGMENTS
We thank U. Surana, O. Cohen-Fix, S. Biggins, K. Hardwick, and M. Lisby for generous gifts of various strains and constructs. We also thank Cheen Fei Chin and Yuan Yuan Chew for technical assistance. We appreciate the insightful comments from the anonymous reviewers. We are grateful to Phuay-Yee Goh, Alice Tay, and Karen Crasta for comments on the manuscript. This work is supported by the BioMedical Research Council (Grant 04/1/21/19/325) and Ministry of Education (grant R183-000-168-112).
Abbreviations used:
- Gal
galactose
- GFP
green fluorescence protein
- MEN
mitotic exit network
- Noc
nocodazole
- Raff
raffinose
- RFP
red fluorescence protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0637) on May 26, 2010.
REFERENCES
- Agarwal R., Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G., Zachariae W., Schleiffer A., Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Amon A. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Baskerville C., Segal M., Reed S. I. The protease activity of yeast separase (esp1) is required for anaphase spindle elongation independently of its role in cleavage of cohesin. Genetics. 2008;178:2361–2372. doi: 10.1534/genetics.107.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Murray A. W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S. Y., Vermaak D., Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Chiroli E., Rancati G., Catusi I., Lucchini G., Piatti S. Cdc14 inhibition by the spindle assembly checkpoint prevents unscheduled centrosome separation in budding yeast. Mol. Biol. Cell. 2009;20:2626–2637. doi: 10.1091/mbc.E08-11-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Elliott S., Knop M., Schlenstedt G., Schiebel E. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA. 1999;96:6205–6210. doi: 10.1073/pnas.96.11.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman V., Gerson-Gurwitz A., Movshovich N., Kupiec M., Gheber L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009;10:387–393. doi: 10.1038/embor.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. K., et al. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S. J., Wheatley E., Rittinger K., Johnston L. H., Sedgwick S. G. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 2002;277:28439–28445. doi: 10.1074/jbc.M202540200. [DOI] [PubMed] [Google Scholar]
- Gillett E. S., Espelin C. W., Sorger P. K. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P. K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E. R., Hoyt M. A. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol. Biol. Cell. 2001;12:3402–3416. doi: 10.1091/mbc.12.11.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L. J., Krutchinsky A. N., Morgan D. O. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa Lim H., Yeong F. M., Surana U. Inactivation of mitotic kinase triggers translocation of MEN components to mother-daughter neck in yeast. Mol. Biol. Cell. 2003;14:4734–4743. doi: 10.1091/mbc.E03-04-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V. B., Murray A. W. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Kang J., Cheeseman I. M., Kallstrom G., Velmurugan S., Barnes G., Chan C. S. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Roostalu J., Roque H., Antony C., Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Krishnan V., Nirantar S., Crasta K., Cheng A. Y., Surana U. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell. 2004;16:687–700. doi: 10.1016/j.molcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee S. E, Frenz L. M., Wells N. J., Johnson A. L., Johnston L. H. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Burke D. J. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- Liu H., Liang F., Jin F., Wang Y. The coordination of centromere replication, spindle formation, and kinetochore-microtubule interaction in budding yeast. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. O. Regulation of the APC and the exit from mitosis. Nat. Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Ross K. E., Cohen-Fix O. The role of Cdh1p in maintaining genomic stability in budding yeast. Genetics. 2003;165:489–503. doi: 10.1093/genetics/165.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M. A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S. C., Liu J. Y., Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Hyman A. A., Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 2001;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 2003;116:4263–4275. doi: 10.1242/jcs.00807. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B., Brown P. O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepel J., Ottey M. A., Kurischko C., Hieter P., Luca F. C. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol. Biol. Cell. 2005;16:5465–5479. doi: 10.1091/mbc.E05-04-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Sedat J. W., Murray A. W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Lehane C., Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D. O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. L., Rida P. C., Surana U. Essential tension and constructive destruction: the spindle checkpoint and its regulatory links with mitotic exit. Biochem. J. 2005;386:1–13. doi: 10.1042/BJ20041415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., Stark M. J., Tanaka K. Kinetochore capture and bi-orientation on the mitotic spindle. Nat. Rev. Mol. Cell Biol. 2005;6:929–942. doi: 10.1038/nrm1764. [DOI] [PubMed] [Google Scholar]
- Thornton B. R., Toczyski D. P. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20:3069–3078. doi: 10.1101/gad.1478306. [DOI] [PubMed] [Google Scholar]
- Tinker-Kulberg R. L., Morgan D. O. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytell J. D., Sorger P. K. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. The mechanism of sister chromatid cohesion. Exp. Cell Res. 2004;296:80–85. doi: 10.1016/j.yexcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A. CDC20 and CDH1, a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Westermann S., Drubin D. G., Barnes G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Wigge P. A., Kilmartin J. V. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., O'Toole E. T. The spindle cycle in budding yeast. Nat. Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- Yeh E., Skibbens R. V., Cheng J. W., Salmon E. D., Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong F. M., Lim H. H., Surana U. MEN, destruction and separation: mechanistic links between mitotic exit and cytokinesis in budding yeast. Bioessays. 2002;24:659–666. doi: 10.1002/bies.10106. [DOI] [PubMed] [Google Scholar]
- Zeng X., Kahana J. A., Silver P. A., Morphew M. K., McIntosh J. R., Fitch I. T., Carbon J., Saunders W. S. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.