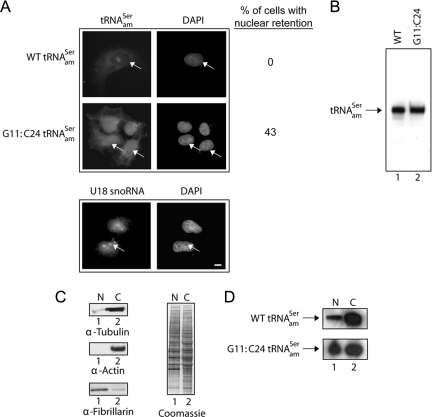
Figure 1.
The G11:C24 mutation affects the efficiency of nuclear export of tRNAamSer. (A) FISH analysis of the nuclear-cytoplasmic distribution of the WT and G11:C24 mutant amber suppressor tRNA. COS-7 cells were transfected with pSVBpUC containing the WT tRNAamSer or G11:C24 tRNAamSer gene. The subcellular distribution of the wild-type tRNAamSer and G11:C24 tRNAamSer mutant and the location of U18 snoRNA were detected using Cy3- and fluorescein-labeled oligonucleotides, respectively. The values presented are averages of three independent experiments. DNA was visualized by staining with DAPI. Bar, 10 μm. (B) The G11:C24 mutation did not affect maturation of tRNAamSer. Total RNA was isolated from COS-7 cells transfected with pSVBpUC carrying the WT (lane 1) or G11:C24 mutant tRNAamSer gene (lane 2). An aliquot containing 20 μg of total RNA was subjected to PAGE under denaturing conditions, and tRNAamSer was detected by Northern blot analysis. (C) Analyses of the purity of nuclei isolated from COS-7 cells. Nuclear (N) and cytoplasmic (C) fractions were prepared from COS-7 cells, and Western blot analysis was used to monitor the amount of the cytoplasmic markers tubulin (left, first row) and actin (second row) associated with nuclei, and the amount of the nucleolar marker fibrillarin present in the cytoplasmic fraction (third row). The amount of protein in the two fractions was assessed by Coomassie Blue staining of the blot (right). (D) Northern blot analysis of the nuclear-cytoplasmic distribution of the WT and G11:C24 mutant tRNAamSer. N and C fractions were isolated from COS-7 cells transfected with pSVBpUC carrying the G11:C24 mutant or WT suppressor tRNA gene. Total RNA as isolated from each fraction and subjected to PAGE under denaturing conditions. The RNA was transferred electrophoretically to membrane, and Northern analysis was performed to detect tRNAamSer.