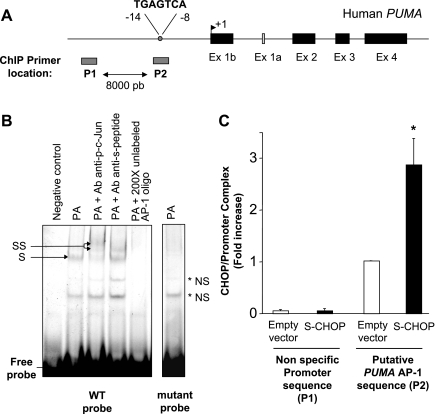
Fig. 4.
S-peptide-tagged CHOP binds to the activator protein (AP)-1 binding site within PUMA promoter. A: genomic structure of PUMA depicting AP-1 binding site within the promoter region. As indicated by the gray bars, P1 and P2 are the two primer sets used for chromatin immunoprecipitation (ChIP) assays, amplifying, respectively, an upstream nonspecific sequence or the AP-1 sequence within the human PUMA promoter. B: Huh-7 cells were transiently transfected for 24 h with an S-peptide-tagged CHOP. Nuclear protein extracts were isolated 8 h after treatment with palmitic acid at 800 μM. EMSA was performed using CY 5.5-labeled AP-1 probe as described in materials and methods. Antibodies against phospho-c-Jun or S-peptide were used in supershift experiments. Retarded complexes are indicated by arrows (S: shift; SS: supershift, NS: nonspecific band). No specific retarded complexes were observed when adding 200-fold molar excess of the unlabeled probe or when using the CY 5.5-labeled mutated AP-1 probe. CY 5.5-labeled AP-1 probe only with no nuclear extract was used as a negative control. C: Huh-7 cells were transiently transfected for 24 h with an S-peptide-tagged CHOP or the empty vector and then treated with palmitic acid at 800 μM for 4 h. Cells were fixed and lysed, and DNA fragments pulled down with S-peptide-tagged CHOP were subjected to quantitative RT-PCR analysis using the primers P1 or P2 as described above. Data represent the means and standard errors of 3 experiments; *P < 0.01, palmitate-treated S-CHOP vs. palmitate-treated empty vector.