Figure 5.
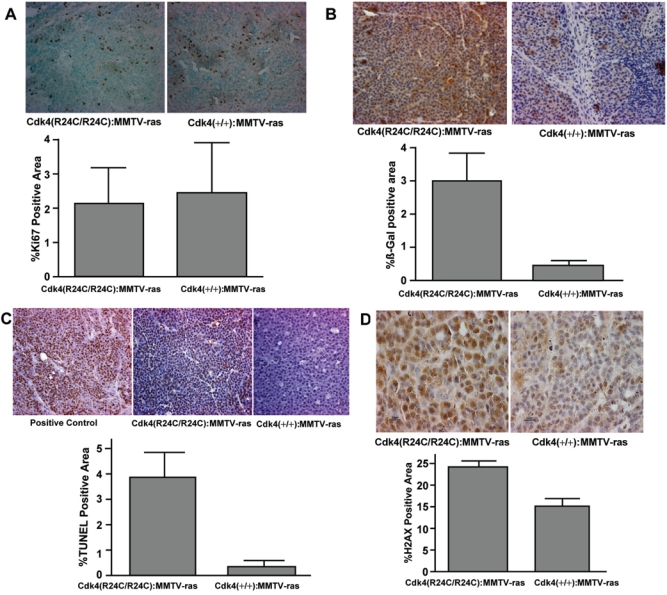
Analysis of tumor sections of Cdk4(+/+):MMTV-v-Ha-ras and Cdk4(R24C/R24C):MMTV-v-Ha-ras mice for proliferation, senescence, apoptosis, and DNA damage markers. (A) Analysis of proliferation marker Ki67 in the tumor sections. (A, top) Formalin-fixed paraffin-embedded normal and tumor sections of the mammary glands of Cdk4(+/+):MMTV-v-Ha-ras and Cdk4(R24C/R24C):MMTV-v-Ha-ras mice were subjected to immunohistochemical analysis using antibody against proliferation marker Ki67 (40x). (A, bottom) Ki67-positive area (dark brown–stained nuclei) of the tumor sections was measured using Bioquant software and analyzed using an unpaired t test provided in Prism software. (B) Increased senescence in the mammary tumor tissues of Cdk4(R24C/R24C):MMTV-v-Ha-ras mice when compared to Cdk4(+/+):MMTV-v-Ha-ras mice. (B, top) Formalin-fixed paraffin-embedded normal and tumor sections of the mammary glands of Cdk4(+/+):MMTV-v-Ha-ras and Cdk4(R24C/R24C):MMTV-v-Ha-ras mice were subjected to immunohistochemical analysis using antibody against β-galactosidase protein (40x). (B, bottom) β-galactosidase positive area (brown-stained area ) of the tumor sections was measured using Bioquant software and analyzed using an unpaired t test provided in Prism software. (C) Mammary tumors of Cdk4(R24C/R24C):MMTV-v-Ha-ras mice exhibit enhanced apoptosis when compared to Cdk4(+/+):MMTV-v-Ha-ras mice. (C, top) Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) analysis of formalin-fixed paraffin-embedded normal and tumor sections of the mammary glands of Cdk4(+/+):MMTV-v-Ha-ras and Cdk4(R24C/R24C):MMTV-v-Ha-ras mice (40x). (C, bottom) TUNEL-positive area of the tumor sections was measured using Bioquant software and analyzed using an unpaired t test provided in Prism software. (D) Increased levels of γ-H2AX in the tumors of Cdk4(R24C/R24C):MMTV-v-Ha-ras mice. (D, top) Immunohistochemical analysis of formalin-fixed paraffin-embedded sections for γ-H2AX (100x). (D, bottom) γ-H2AX-positive area of the tumor sections was measured using Bioquant software and analyzed using an unpaired t test provided in Prism software.
