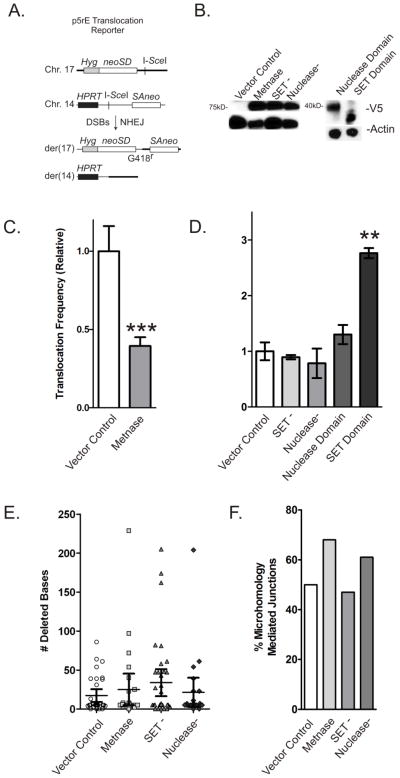
Figure 2. Metnase suppresses chromosomal translocations.
(A) Dual DSB translocation assay in p5rE mES cells as described by Weinstock et al. [4]. I-SceI induces DSBs near splice-acceptor and -donor (SA, SD) sequences attached to truncated neo genes on chromosomes 14 and 17 that become linked by translocation producing a functional neo gene. (B) Expression of V5-tagged Metnase and Metnase mutants and domains (top row) and actin control (bottom row) in p5rE mES cells. (C) Translocation frequencies in mES cells expressing wild-type Metnase are shown relative to cells with empty vector control. Values are averages (+ SEM) for 3 determinations. *** indicates P < 0.0005 (t test). (D) Similar to C, but cells expressing mutant Metnase or domains as labeled. Values are averages (+ SEM) for 3 determinations. * indicates P < 0.05, ** indicates P < 0.01 (t test). (E) Average deletion size among 23–36 products per cell type; no statistically significant differences. (F) Percentage of products with microhomology mediated re-joining in each cell type; no statistically significant differences.