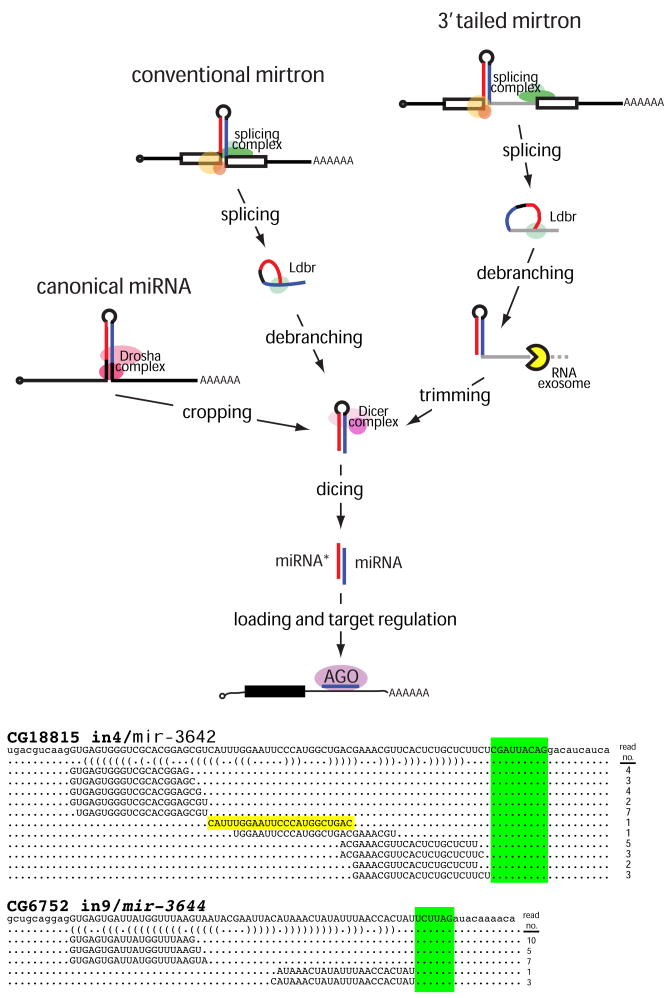
Figure 4.
Pathways that generate miRNA-class regulatory RNAs from short hairpins in Drosophila. In the canonical pathway, the RNase III enzyme Drosha “crops” a primary miRNA transcript to release the pre-miRNA. In the conventional mirtron pathway, the splicing complex liberates short hairpin introns from protein-coding genes. Following their linearization by lariat debranching enzyme (Ldbr), these can fold into pre-miRNA mimics. In the 3′ tailed mirtron pathway, a 3′ tail separates the hairpin from the 3′ splice acceptor site. These require Ldbr and the RNA exosome to remove the 3′ tail up to the hairpin, yielding the pre-miRNA. In all cases, pre-miRNA hairpins are cleaved by the cytoplasmic RNase III Dicer-1 to yield miRNA/miRNA* duplexes. One strand is predominantly selected for incorporation into an AGO1 complex to repress seed-matched targets. At the bottom are additional examples of D. melanogaster tailed mirtrons whose inferred trimmed tails are highlighted green (see also Supplementary Figure 6). Note that the precise terminal loop of the CG18815_in4/mir-3642 tailed mirtron was also cloned (yellow), providing additional evidence for Dicer cleavage.