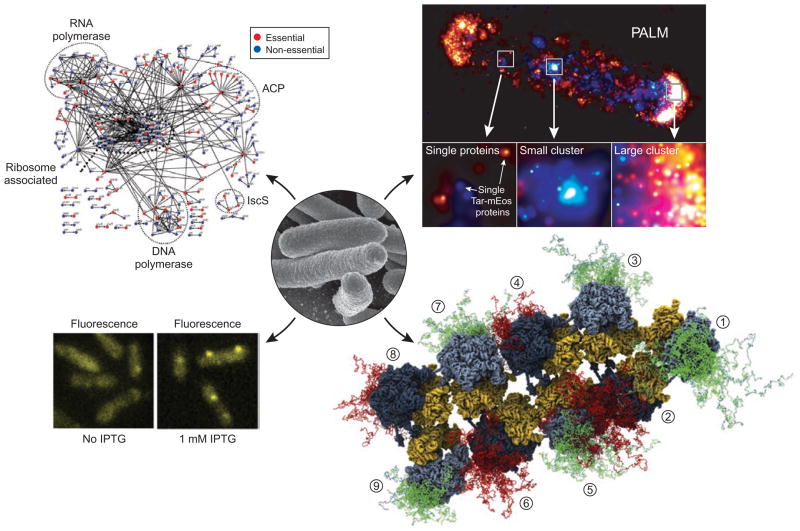
Figure 2.
Examples of powerful new methods to study proteins in their native environments, networks and complexes. A scanning electron micrograph of E. coli cells is shown in the center (from the US National Institute of Allergy and Infectious Diseases; http://www3.niaid.nih.gov/topics/biodefenserelated/biodefense/publicmedia/image_library.html). Clockwise around the cells are shown, beginning with the upper left: an E. coli protein interaction network35; an image of the chemotaxis receptors of E. coli obtained using subdiffraction fluorescence microscopy (photoactivation light microscopy, PALM)33; a reconstructed schematic of E. coli polysomes obtained by fitting ribosomal structures to cryo-electron tomography images and modeling the nascent chains (shown in green or red) emerging from individual ribosomes (numbered from 1 to 8)15; and localization of T7 RNA polymerase molecules (labeled with a fluorescent protein marker) to promoters on DNA upon IPTG induction of transcription31.