Figure 1. Map-Shuffle-Scan framework used by Crossbow.
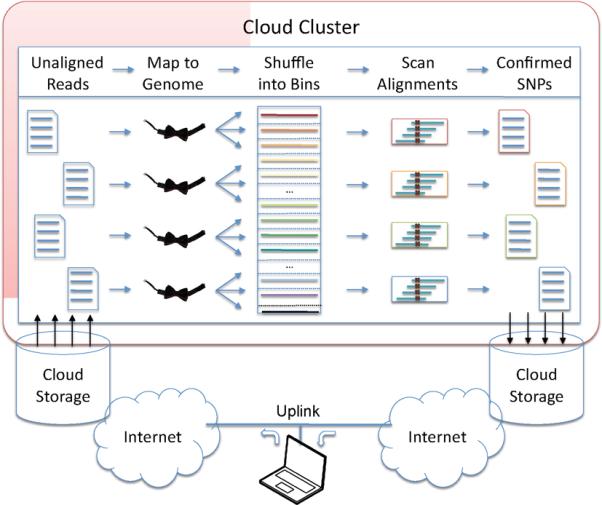
Users begin by uploading the sequencing reads into the cloud storage. Hadoop, running on a cluster of virtual machines in the cloud, then maps the unaligned reads to the reference genome using many parallel instances of Bowtie. Hadoop then automatically shuffles the alignments into sorted bins determined by chromosome region. Finally, many parallel instances of SOAPsnp scan the sorted alignments in each bin. The final output is a stream of SNP calls stored within the cloud that can be downloaded back to the user's local computer.
