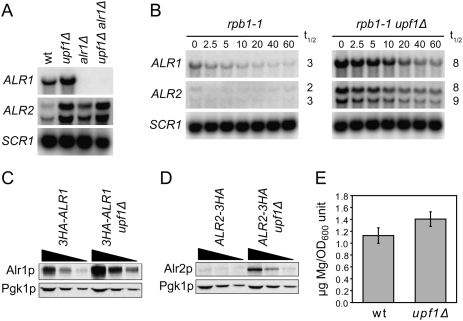
Figure 2.
NMD-deficient cells show increased expression of magnesium transporters. (A) Northern analysis of total RNA isolated from wild-type (MJY142), upf1Δ (MJY67), alr1Δ (MJY130), and upf1Δ alr1Δ (MJY447) cells grown in SC medium at 30°C. The blot was probed for ALR1, ALR2, and SCR1 transcripts using randomly labeled DNA fragments. (B) mRNA decay rates in rpb1-1 (MJY249) and rpb1-1 upf1Δ (MJY251) cells. The strains were grown at 24°C in SC medium followed by inhibition of RNA polymerase II transcription by a shift to 37°C. Time points (minutes) after the shift are indicated above the lanes. The signal in each lane was normalized to the corresponding SCR1 signal, and the half-life (t1/2, in minutes) was determined from the initial slope of the curve. (C,D) Western blot analyses of twofold serial dilutions of cell extracts from strains in which the DNA sequence for three tandem influenza virus hemagglutinin epitopes (3HA) was fused to the endogenous ALR1 (C, MJY290 and MJY309) or ALR2 (D, MJY167 and MJY183) ORF. Monoclonal antibodies against HA or Pgk1p were used to detect the indicated proteins. Control experiments showed that the sequence for the 3HA tag did not prevent accumulation of the transcripts in upf1Δ cells (Supplemental Fig. S2). (E) Mg2+ levels in supernatants of permeabilized wild-type (MJY142) and upf1Δ (MJY67) cells. The values, determined by using eriochrome blue SE, represent the average from four independent cultures of each strain. The standard deviation is indicated. A two-tailed Student's t-test revealed that the value for the upf1Δ strain is significantly different from that of the wild type (P = 0.02).