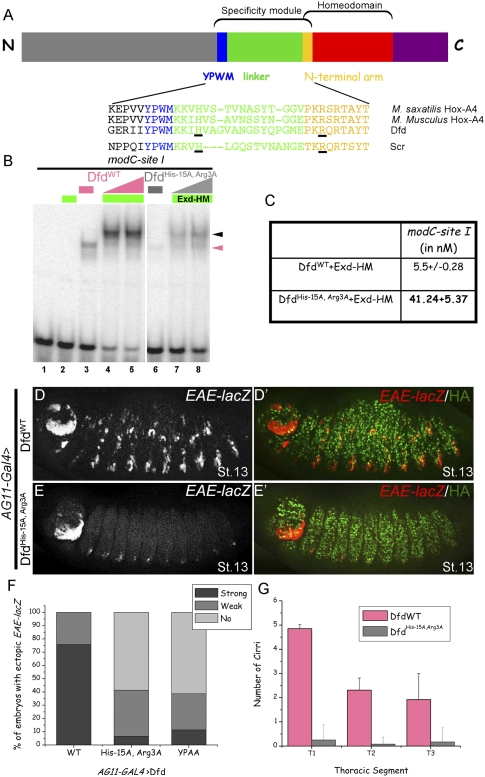
Figure 3.
Dfd specificity module residues His-15 and Arg3 are required in vitro and in vivo for Dfd-specific functions. (A) Schematic showing Hox specificity module, homeodomain, YPWM motif, and linker. Below are alignments of specificity modules from Dfd orthologs, showing the conserved histidine and an arginine (underlined) at positions analogous to those in Scr's specificity module (Joshi et al. 2007). (B) DfdHis-15A, Arg3A binding to site I is weaker both as a monomer (lane 6) and as cooperative trimer (lanes 7,8) with Exd–HM in comparison with DfdWT (lanes 3–5). Hox monomer and Hox–Exd–HM trimer complexes are indicated by pink and black arrowheads, respectively. (C) Kd measurements for DfdWT–Exd–HM and DfdHis-15A, Arg3A–Exd–HM to site I show an eightfold difference in binding affinity. (D,E) Ubiquitous expression of DfdWT results in ectopic activation of EAE-lacZ (D), while DfdHis-15A, Arg3A (E) is severely compromised in its ability to induce this reporter gene (see F for quantification). (F) Graph showing percentage of total embryos that activate EAE-lacZ ectopically when DfdWT, DfdHis-15A, Arg3A, or DfdYPAA is expressed ubiquitously using AG11-Gal4 (n ≥ 56 for each genotype, n = 2). (G) Graph showing the quantification of ectopic cirri made in thoracic segments of embryonic cuticles when DfdWT or DfdHis-15A, Arg3A is expressed ubiquitously using AG11-Gal4. Error bars represent standard deviations.