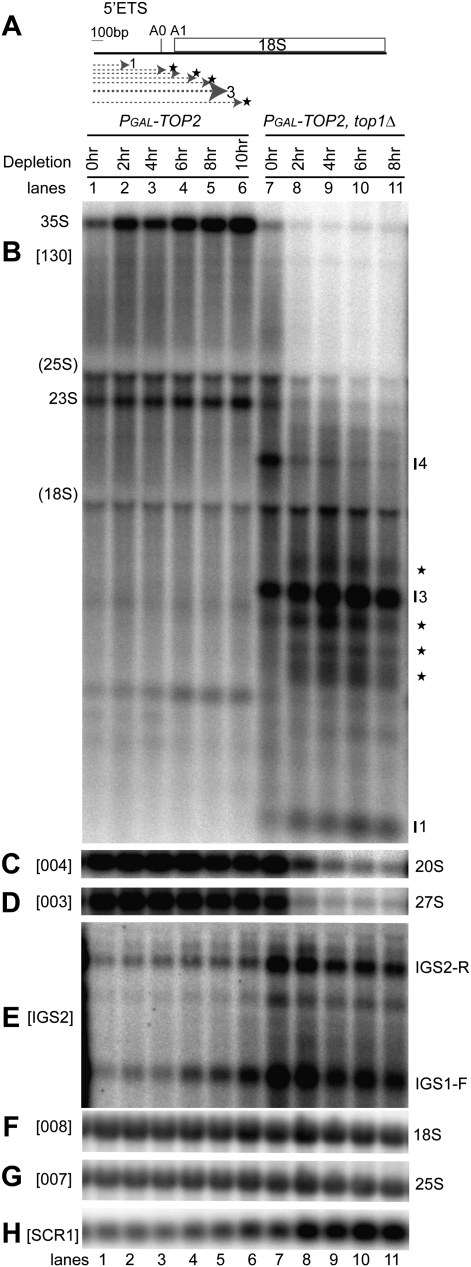
Figure 3.
Pre-rRNAs are truncated predominately within the 5′ region of 18S rRNA when both Top1 and Top2 are absent. (A) Diagram representing the 5′-ETS and 18S rDNA sequences. Heterogeneous, truncated pre-rRNA fragments that accumulate in the absence of both Top1 and Top2 are depicted by dotted arrows followed by a star. (B–H) Strains PGAL-TOP2 and PGAL-TOP2 top1Δ were shifted from galactose-containing medium (0 h) to glucose-containing medium at 30°C (2–10 h). Total RNA was analyzed by Northern hybridization. The membrane was hybridized successively with probes 130 (B), 004 (C), 003 (D), 008 (F), and 007 (G). (E) Random primed probe IGS2 was used to detect ncRNAs transcribed by RNA Pol II from the intergenic rDNA spacers IGS1 and IGS2, located between the 35S rDNA units. (H) Cytoplasmic SCR1 RNA was used as a loading control. Intact pre-rRNAs and truncated pre-rRNA fragments are labeled. Heterogeneous truncated pre-rRNA fragments are labeled by stars. “(18S)” and “(25S)” indicate the position of migration of 18S and 25S rRNAs. Probe names are bracketed (see Fig. 1A for location of probes).