Figure 4.
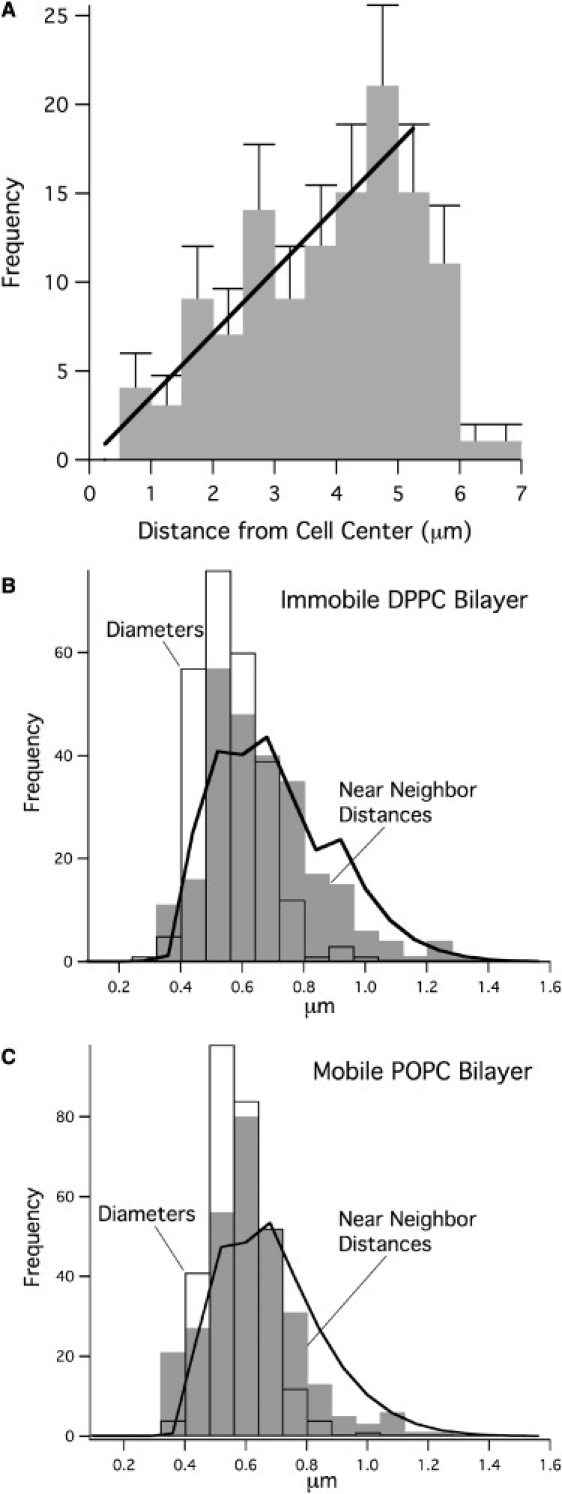
(A) The number of clusters at a radial distance r is proportional to the distance from the center of a cell (Fig. 3A) as expected for a uniform density of randomly positioned clusters. Distributions of cluster diameters and nearest-neighbor distances on immobile (B) and mobile (C) ligand substrates. The area of each bar is the fraction of clusters with nearest-neighbor distances (gray bars) or diameters (white bars) in the x-range of the bar. The solid lines are the nearest-neighbor distances expected from a random spot model P(r) ∝ r exp(−ρπr2), where ρ is the measured density of clusters, weighted with the measured distribution of cluster diameters (see Mathematical Background). Cluster density = 1.139 clusters/μm2 on DPPC (255 clusters on 3 cells), and 1.145 clusters/μm2 on POPC (296 clusters on 3 cells). On immobile DPPC (B), the nearest-neighbor distribution predicted for random clusters fits the observed distribution well. On mobile POPC (C), the fit is slightly poorer, with clusters appearing closer together than predicted for a perfectly random distribution. This may reflect the initial stages of membrane reorganization that lead to the formation of the large patch or synapse on mobile substrates.
