Fig. 1.
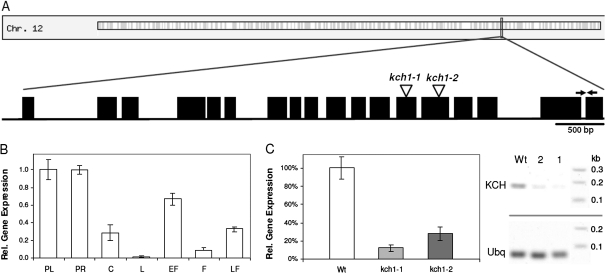
Expression pattern of OsKCH1 in rice and Tos17 insertion mutants. (A) Schematic representation of the OsKCH1 gene structure. Introns are shown as lines, exons as boxes. Positions of Tos17 insertion sites in kch1-1 and kch1-2 are indicated by arrow heads. Positions of the primers used for gene expression analysis are indicated by arrows. (B) Expression of OsKCH1 in rice seedlings and adult plants of the wild type measured by real-time PCR. PL, primary leaf; PR, primary root; C, coleoptile; L, adult leaf; EF, young flower bud; F, fully developed fower; LF, flower after pollination. (C) Expression of OsKCH1 in Tos17 insertion mutants quantified by real-time PCR in RNA extracts from complete seedlings of the mutant lines kch1-1 and kch1-2 as compared to the wild-type control. In addition, a representative RT-PCR shows the abundance of OsKCH transcripts in seedlings of the wild type as compared to the mutants kch1-1 (1) and kch1-2 (2). Ubiquitin (Ubq) was used as internal standard in all samples.