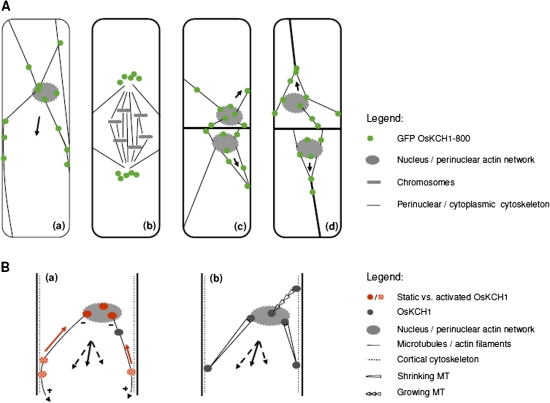
Fig. 8.
OsKCH1 localization throughout the cell cycle and working models for OsKCH1 function in nuclear positioning. (A) Dynamic redistribution of GFP OsKCH1 during the cell cycle. In interphasic and premitotic cells (a), OsKCH1 signals colocalized with a cytoskeletal network that tethers the nucleus to the periphery. Punctate signals accumulated at the sites, where the filaments reached the nuclear envelope or connected to the cell cortex. During mitosis (b), OsKCH1 retracted from nucleus and division plane and did not colocalize with mitotic microtubule structures. During cytokinesis, OsKCH1 signals were repartitioned and subsequently mainly found surrounding the newly formed nuclei, again along the cytoskeletal network that tethered these nuclei to the periphery or connected the nuclei with the cell poles. Arrows indicate the observed directions of nuclear movement. (B) Schematic models for the function of OsKCH1 in nuclear positioning. A ‘sliding model’ (a) is based on two different populations of OsKCH1. Static OsKCH1 proteins anchor the minus (–) end of radial microtubules at the nuclear envelope, possibly via interaction with the perinuclear actin network. The plus (+) ends of the microtubules are captured at the cortex by protein complexes involving activated OsKCH1 motors that move towards microtubule minus-ends (red arrow), but are anchored such that they generate a sliding force (arrowheads) that finally acts on the nucleus. Black arrows depict the force vectors that produce a resulting force (plain arrow) pointing in the observed directions of nuclear movement. A ‘pulling/pushing model’ (b) relies rather on microtubule dynamics. Microtubules are captured by anchor protein complexes involving OsKCH1 on both the nuclear envelop and the cell cortex. Depending on the accessory protein complexes, microtubule dynamics are differentially regulated, resulting either in growth or shrinkage. Black arrows again depict the force vectors. (This figure is available in colour at JXB online.)