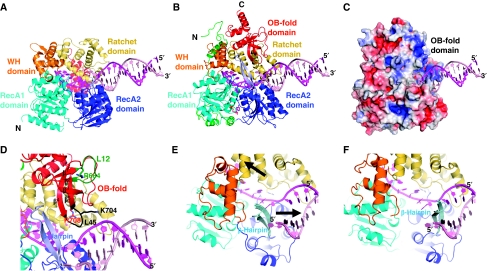
Figure 3.
RNA unwinding by the Prp43p helicase. (A) Overview of the Hel308/DNA complex (PDB 2P6R; Buttner et al, 2007) in the same orientation and colour code as in Figure 1B. The DNA strands are shown in magenta (threaded strand) and pink cartoons. (B) Model of the Prp43p/DNA complex modelled against the Hel308/DNA complex by simple superposition of the proteins (same orientation and colour code as in (A)). (C) Surface representation of Prp43p in complex with modelled DNA (magenta and pink), coloured according to its electrostatic potential calculated with APBS (Baker et al, 2001) and presented in the same orientation as in (B). (D) Close-up view of the nucleic acid-binding cavity entrance of the Prp43p/DNA complex. The β-hairpin element and the OB-fold domain are coloured light blue and red, respectively. The loops L12 (dark green) and L45 (black) and specific positively charged residues (pictured as sticks) of the OB-fold, which are mutated in the probed Prp43p variants, are also indicated. (E, F) Close-up views of the conformational rearrangements of each of the Prp43p domains (E, initial conformation) to adopt the Hel308 conformation (F, final conformation). The Prp43p domains and modelled DNA are pictured as in (B). The two black arrows in (E) show the concerted movement of the RecA-like and ratchet domains, which provides a possible model for RNA translocation and unwinding mechanism of Prp43p helicase: the ratchet domain seems to pull the threaded single strand inside the cavity (left arrow), whereas the β-hairpin slices through the nucleic acid duplex (right arrow).