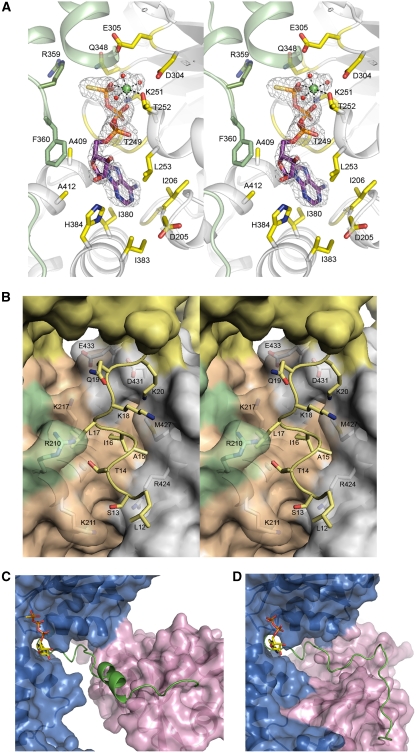
Figure 3.
Changes in the structure of N–D1 on binding of ATPγS. (A) Stereoscopic pair showing details of the ATPγS-binding environment. The nucleotide-binding site is located at the subunit interface. One subunit is coloured in green and the other in grey. Residues making contact with bound ATPγS are drawn as sticks and are labelled. The ATPγS molecule is shown as a stick model with carbon in purple, oxygen in red, nitrogen in blue, phosphorous in magenta, and sulphur in yellow. The ATPγS molecule is enclosed in a difference electron density cage in grey, contoured at the 2.5σ level. The Mg2+ ion is shown as a green ball with the three coordinating water molecules in red. (B) Stereo pair showing the ordering of an N-terminal segment from Leu12 to Lys20 in the ATPγS form and its environment. The re-ordered residues are depicted as stick models in yellow and their environment is shown as a semi-transparent molecular surface with all interacting residues rendered as stick models with labels. The yellow surface is the N-domain; orange and green surfaces are the cognate D1-domain and N–D1 linker, respectively. The grey surface is from an adjacent D1-domain. (C) Conformation of the N–D1 linker in the presence of ATPγS. The N–D1 linker is shown as a ribbon model in green; N- and D1-domains are shown as molecular surfaces in magenta and in blue, respectively. The bound ATPγS is shown as a stick model. (D) Conformation of the N–D1 linker in the presence of ADP. The N–D1 linker is shown as a coil in green. The D1-domain is similarly orientated as in (C). The bound ADP is shown as a stick model.