Figure 1. Bi-domain structure of histamine N-methyltransferase.
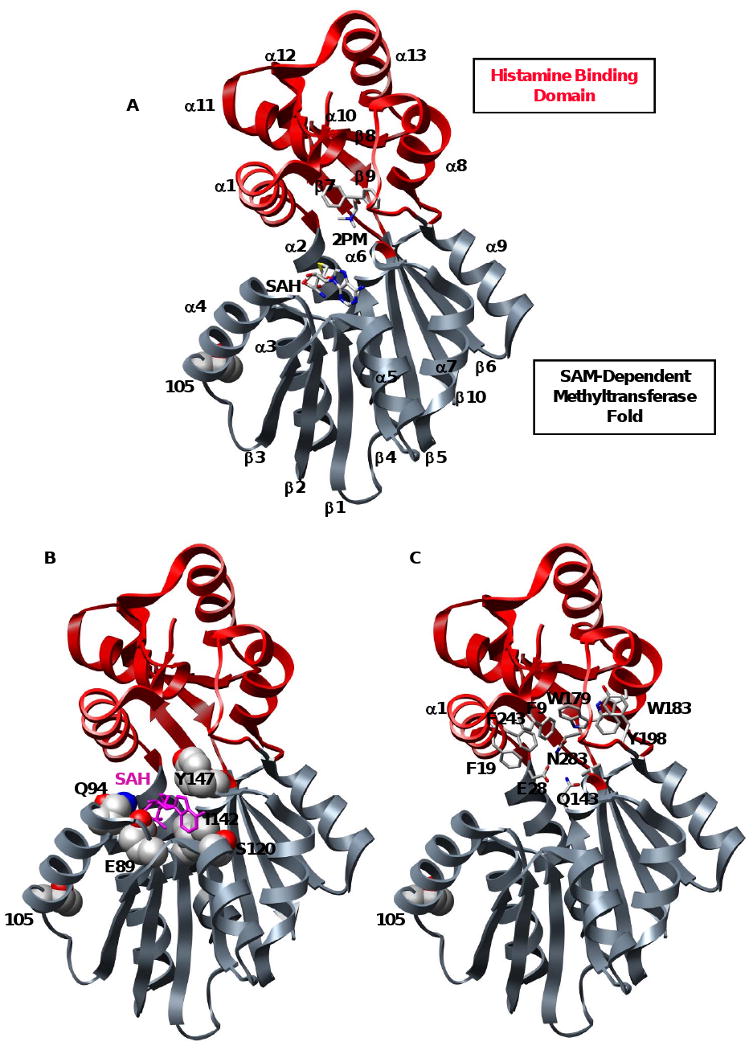
(A) Ribbon diagram of HNMT (2AOT) with the SAM and histamine binding domains colored gray and red, respectively. Product S-adenosylhomocysteine (SAH), inhibitor diphenhydramine (2PM), and polymorphic residue 105 are colored by atom. (B) Key residues in the highly conserved SAM-dependent methyltransferase domain. SAM interacts with residues from α4 (Q94), α5 (S120), β2 (E89, P90), β3 (T119) and β4 (I142, M144). SAH is colored magenta and shown in licorice representation, residues are colored by atom and shown in space-filling representation. (C) Key residues in the histamine-binding domain. A trio of catalytic residues (E28, Q143 and N283) is involved in the N-methylation of HNMT substrates. The substrate is buried in a hydrophobic domain lined with aromatic residues (shown in licorice representation).
