Figure 5. Snapshots of polymorphic packing taken from the 20 ns structures of the 105T and 105I HNMT MD simulations at 37°C.
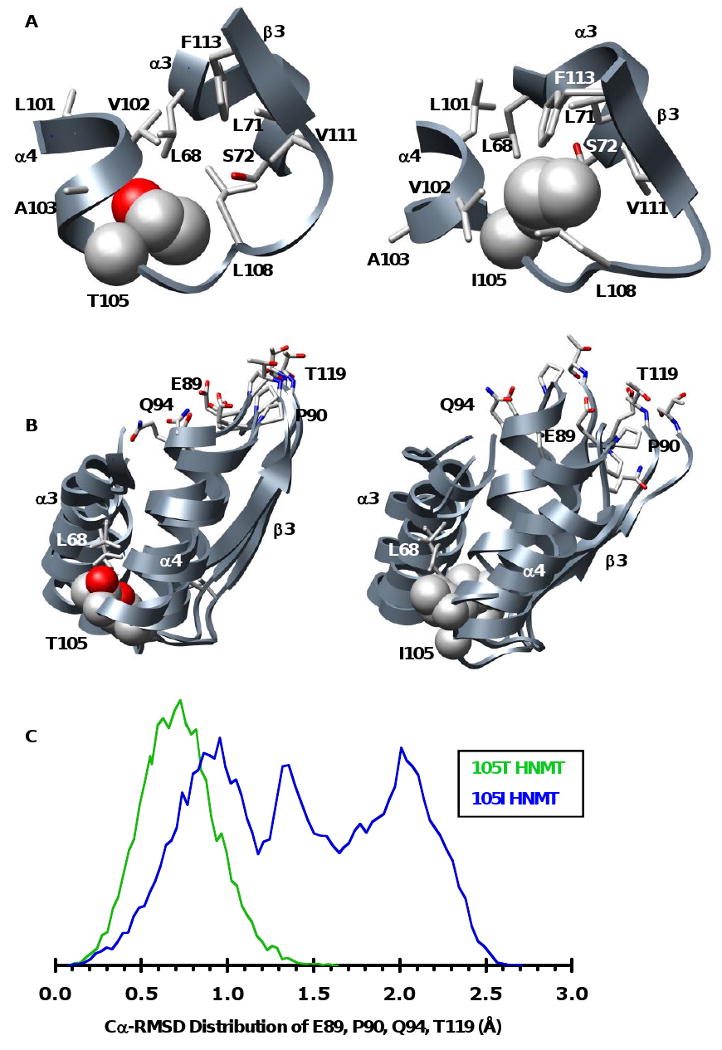
(A) Packing at the polymorphic site. The larger Ile is more buried, forming side-chain contacts with residues from α3 (L71, S72) and β3 (V111, F113) that are not present in the 105T simulations. (B) The polymorphic packing affects the SAM-binding site. Structural overlay of the polymorphic and SAM binding-sites from the 20ns structures of three independent simulations of 105T and 105I HNMT. The more tightly packed Ile acts as a pivot point between α3, α4 and β3 disrupting the orientation of key SAM-binding residues (E89, P90, Q94, T119). The side chain of the polymorphic residue and its contacts are colored by atom and shown in space-filling and licorice representations, respectively. (C) Distributions of the Cα-RMSDs for residues E89, P90, Q94 and T119 during the last 10ns of all three of the 105T (green) and 105I (blue) HNMT simulations at 37°C. The SAM-binding residues are more mobile in 105I HNMT, existing in a large ensemble of conformations that differ greatly from their respective positions in the starting structure. The distribution for 105T HNMT is narrower, indicating that the active site structure of 105T HNMT is maintained throughout the simulations. The vertical scales are arbitrary.
