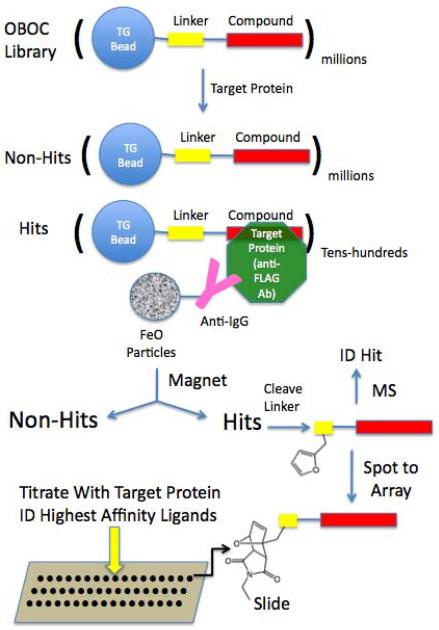
Figure 1. Overview of the integrated magnetic screening and testing of hits on microarrays.
Millions of 75 μm TentaGel beads from a one bead one compound (OBOC) library are incubated with target protein (anti-FLAG antibody in this study), washed, then incubated with anti-target protein antibodies linked covalently to iron oxide containing particles (Dynabeads). Beads that bind the target protein and therefore also attract Dynabeads, are retained on the side of the tube using a powerful magnet and non-magnetic beads are removed. Each of the putative “hit” beads is separated into the well of a microtiter plate and the compounds are removed from the beads by cleavage of a linker. The compounds are then spotted onto a maleimide-activated glass slide via a Diels-Alder reaction involving a conserved furan-containing monomer incorporated into each sequence. The structure of each putative hit is deduced by tandem mass spectrometry (MS). The compound microarrays are then probed with different concentrations of the target protein to determine the intrinsic affinity of each of the hit compounds for the target. In this way, no re-synthesis of the hits is necessary until the best binders are identified.