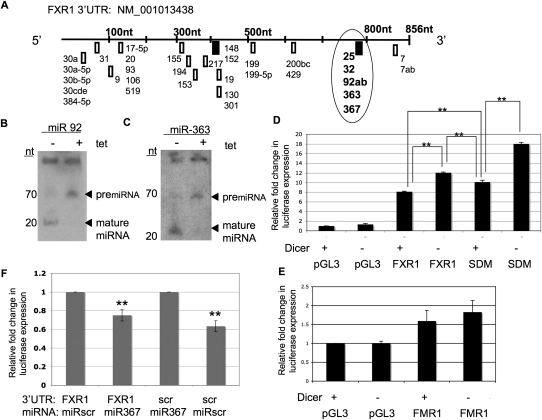
FIGURE 3.
FXR1 3′UTR contains a common seed sequence that confers miRNA-mediated regulation on luciferase constructs. (A) Schematic of the predicted miRNA seed sequence binding sites in the FXR1 3′UTR based on TargetScan 5.1. miRNAs are labeled. (Boxes) Seed sequences, (bold boxes) the two 8-nt sites with higher probability of preferred conservation, (circle) location of the 8-nt seed sequence common to miRNAs-25/32/92/363/367. miR-7 was predicted by TargetScan 5.1 but was not included in the FXR1 3′UTR subcloned luciferase constructs. (B,C) Total RNA (50 μg) from DT40 cells treated with tetracycline for 96 h (+) or mock treated (−) was resolved on a 15% acrylamide gel and probed with [32P]-labeled antisense oligos to miR-92 (B) and miR-363 (C), as indicated (top). (Left) Size in nucleotides, (right) positions of pre-miRNAs (upper band) and mature miRNAs (lower band). (D) pGL3 luciferase reporter vector alone (pGL3), containing the 3′UTR of FXR1 (FXR1), or containing the FXR1 3′UTR in which the seed sequence common to miRNAs 25/32/92/363/367 was deleted (SDM) were electroporated into DT40 cells after 72 h growth in tetracycline. Twenty-four hours after electroporation (for 96 total hours in tetracycline), luciferase expression was assessed in DT40 cells grown in the presence (+) or absence (−) of tetracycline (tet). Transfection efficiency was monitored by Renilla expression, and all luciferase values were normalized to empty vector (pGL3) in the absence of tetracycline. Values are depicted as expression fold change compared with empty vector. Significance was determined by Student's t-test. (**) P-value < 0.007, n = 3. (E) pGL3 vector containing the FMR1 3′UTR was electroporated into DT40 cells under the conditions described above. No significant difference in luciferase expression was observed in the presence (−) or absence (+) of miRNAs by Student's t-test (P-value = 0.392, n = 3). (F) pGL3 vector containing the FXR1 3′UTR or the FXR1 3′UTR in which the 25/32/92/363/367 was deleted and replaced with the sequence CCTGTTAG (scr) were introduced into HEK293 cells with either the miR367 mimic or the miRNA complementary to the CCTGTTAG (scr) sequence (miR-scr), as indicated below. Luciferase and Renilla activity was measured and ratios calculated. Values are depicted as expression fold change determined by comparing luciferase expression in the presence of the complementary miRNA to luciferase expression in the presence of the opposite, nonbinding miRNA, the latter value designated as 1. Relative fold change is indicated. Significance was determined by Student's t-test. (**) P-value < 0.01, n = 3.