Abstract
The fly brain is formed by approximately hundred paired lineages of neurons, each lineage derived from one neuroblast. Embryonic neuroblasts undergo a small number of divisions and produce the primary neurons that form the functioning larval brain. In the larva, neuroblasts produce the secondary lineages that make up the bulk of the adult brain. Axons of a given secondary lineage fasciculate with each other and form a discrete bundle, the secondary axon tract (SAT). Secondary axon tracts prefigure the long axon connections of the adult brain, and therefore pathway choices of SATs made in the larva determine adult brain circuitry. Drosophila Shotgun/E-cadherin (DE-cad) and its binding partner Armadillo/β-catenin (β-cat) are expressed in newly born secondary neurons and their axons. The fact that the highly diverse, yet invariant pattern of secondary lineages and SATs has been recently mapped in the wild-type brain enabled us to investigate the role of DE-cad and β-cat with the help of MARCM clones. Clones were validated by their absence of DE-cad immuno-reactivity. The most significant phenotype consists in the defasciculation and an increased amount of branching of SATs at the neuropile-cortex boundary, as well as subtle changes in the trajectory of SATs within the neuropile. In general, only a fraction of mutant clones in a given lineage showed structural abnormalities. Furthermore, although they all globally express DE-cad and β-cat, lineages differ in their requirement for DE-cad function. Some lineages never showed morphological abnormalities in MARCM clones, whereas others reacted with abnormal branching and changes in SAT trajectory at a high frequency. We conclude that DE-cad/β-cat form part of the mechanism that control branching and trajectory of axon tracts in the larval brain.
Keywords: DE-Cadherin, Drosophila, brain, lineage, pathfinding
Introduction
Neurons are morphologically characterized by long, highly branched cytoplasmic extensions called neurites (dendrites and axons). Axonal pathfinding and the pattern of neurite branching determines the wiring of neuronal circuits. Thus, guiding axons along a specific pathway, or setting a branchpoint at one place of the neurite versus another will alter significantly the way in which that neuron is connected to other neurons. The developing brain of Drosophila has proven to be an excellent model system to unravel the genetic mechanisms that control pathway choices and branching behavior of neurons.
The Drosophila brain is formed by a stereotyped set of approximately 100 neuroblasts that appear in the early embryo (Urbach and Technau, 2003; Younossi-Hartenstein et al., 1996). Each neuroblast produces a small lineage of primary neurons during the embryonic period. Neurons that belong to one lineage remain clustered together; likewise, their axons form a coherent bundle, the primary axon tract (PAT). Primary axons then elaborate axonal and dendritic arbors which establish the neuropile of the larval brain. After a period of mitotic quiescence that lasts from mid-embryogenesis to mid larval development, neuroblasts resume their activity and produce much larger lineages of secondary neurons. Similar to primary axons, axons of a given secondary lineage fasciculate with each other and form a discrete bundle, the secondary axon tract (SAT) within the larval brain (Dumstrei et al., 2003b; Pereanu and Hartenstein, 2006). SATs most often remain a single, undivided tract as they enter the neuropile; in certain lineages, the SAT splits into two or even three branches at the cortex-neuropile boundary, and these SAT branches travel along separate pathways in the neuropile to connect to specific compartments. The pathways defined by the SATs in the larval brain define long axon connection of the adult brain (Dumstrei et al., 2003b; Nassif et al., 2003; Pereanu and Hartenstein, 2006; Pereanu et al., 2009). The only features added to secondary neurons during metamorphosis are the proximal and terminal branches that represent sites of postsynaptic input and presynaptic output. It follows that pathway choices of SATs made during the larval stage determine the “macrocircuitry”, that is, the pattern in which brain compartments are connected. To investigate the mechanisms underlying circuitry, it therefore seems appropriate to study the impact of genes on the pathway choices of lineages made at the larval stage. In this paper we have used the MARCM technique to analyze the role of Drosophila E-cadherin (DE-cad) and its binding partner, Armadillo/β-catenin (β-cat) in the formarion of SATs in the larval brain.
The cadherins form a family of widely expressed adhesion molecules that, according to many studies in vertebrates and invertebrates alike, (Hirano et al., 2003; Tepass et al., 2000) play a central role in patterning of neuronal connectivity. Cadherins consist of an extracellular domain with tandem cadherin repeats, a single membrane-spanning segment, and a cytoplasmic region (Hill et al., 2001; Nollet et al., 2000; Tepass et al., 2000). The so-called classical cadherins interact in a dynamic manner with the actin cytoskeleton via binding to a complex of cytoplasmic proteins, the catenins, The Drosophila genome contains two well studied classical cadherins, DE-cad and DN-cadherin (DN-cad) (Hill et al., 2001; Tepass et al., 2000).. DN-cad is expressed in differentiating neurons and is mainly involved in late events of neuronal development, in particular the formation and maintenance of synaptic connections (Hummel and Zipursky, 2004; Iwai et al., 2002; Prakash et al., 2005). Interfering with DN-cad function disrupts axon-target interaction in the optic lobe and antennal lobe-to-mushroom body projection (Lee et al., 2001; Zhu and Luo, 2004). DE-cad is expressed and required at early developmental stages in epithelial cells; it is downregulated in non-epithelial cells of embryos, including the central nervous system (Oda et al., 1994; Tepass et al., 1996). By contrast, high levels of DE-cad appear postembryonically in the brain where expression can be seen transiently in neuroblasts and secondary neurons, as well as in glia and trachea (Dumstrei et al., 2003b). Knock-down experiments of DE-cad (by expression of a dominant negative construct) revealed a complex larval brain phenotype, consisting in defects of neuroblast proliferation, as well as neuronal and glial morphogenesis (Dumstrei et al., 2003a; Wang et al., 2004). To elucidate the role of DE-cad in a more detailed manner, we decided to undertake an analysis of homozygous loss-of-function clones in the background of a heterozygous (i.e., functionally wild-type) brain. This approach is necessitated by the fact that flies homozygous for loss-of-function alleles in DE-cad or β-cat (as well as most other genes with essential function in neuronal development) are embryonically lethal, and can therefore not be studied at later developmental stages
To generate labeled mutant we used the MARCM technique (Lee and Luo, 1999) in which somatic recombination removes an inhibitory Gal80 construct, which then allows Gal4 driven GFP expression in a clone. The clonal strategy has been widely used in the imaginal discs (eye, wing) and has yielded decisive insight into gene function during pattern formation. It should be pointed out that mutant clones can only be interpreted if the corresponding wild-type clones are well characterized. This has been the case for structures like the eye disc or wing disc epithelium, but not the brain, where every clone (i.e., every lineage) has a different morphology. We have recently mapped and characterized all lineages of the larval brain (Pereanu and Hartenstein, 2006). This map enables us to recognize the identity of individually labeled lineages, be it wild type, or mutant, which, for the first time, makes the analysis of brain clones, induced by somatic recombination, a feasible task. Our data show that DE-cad and β-cat are involved in pathway choices of SATs. Mutant clones had extraneous branches that embarked upon abnormal trajectories in the neuropile. There seemed to be significant differences in the requirement for DE-cad function; some lineages showed no morphological abnormality in MARCM clones, whereas others reacted with abnormal branching at a relatively high frequency. This finding emphasizes the need to assay many lineages when conducting genetic studies of neural development; different lineages/neurons will often differ in their requirement for a given gene.
Materials and Methods
Fly stocks and generation of larval brain clones
Oregon R flies were used as wild-type stock. The following lines were requested from the Bloomington stock center: (1) hs-FLP, elav-Gal4, UAS-mCD8-GFP/FM7c; (2)hs-FLP, elav-Gal4, UAS-mCD8-GFP; FRT42D; (3) FRT42B, UAS-GFP/CyO; (4) FRT19A; (5) y, w*, arm4/FM7c; (6) y, arm1/Fm7c; (7) w*, hs-FLP, FRT19A tub-Gal80; (8) elav-gal4; and (9)chaT-gal4, UAS-GFP. FRT42B, shgR69, UAS-GFP/CyO and FRT42B, hsπmyc, tub-gal80/CyO; Tub-Gal4/TM6B were provided by Dr. Carthew (Hayashi and Carthew, 2004).
To generate DE-cad mutant clones using the MARCM technique, two different mating schemes were used. (1) males of hs-FLP, elav-Gal4, UAS-mCD8-GFP/FM7c; FRT42D tub-Gal80 were crossed with females of hs-FLP, elav-Gal4, UAS-mCD8-GFP; FRT42D shgR69/CyO flies. (2) males of hs-FLP; FRT42B, shgR69, UAS-GFP/+ were crossed with females of FRT42B, hsπmyc, tub-gal80/CyO; elav-gal4/TM6B (Hayashi and Carthew, 2004). To generate comparable wild-type clones, two different mating schemes were used. (1) males of hs-FLP, elav-Gal4, UAS-mCD8-GFP/FM7c; FRT42D Tub-Gal80 were crossed with female hs-FLP, elav-Gal4, UAS-mCD8-GFP; FRT42D/CyO flies. (2) males of hs-FLP; FRT24B, UAS-GFP/+ were crossed with FRT42B, hsπmyc, tub-gal80/CyO; elav-gal4/TM6B.
To generate arm mutant clones, males of hs-FLP, FRT19A tub-Gal80; elav-Gal4, UAS-GFP/UAS-GFP were crossed with females of FRT19A arm1/FM7c flies. Control clones were generated by crossing males of hs-FLP, FRT19A, Tub-Gal80; elav-Gal4, UAS-GFP/UAS-GFP with females of FRT19A/FRT19A.
Embryos were collected on apple juice plates for 1 hour at room temperature. Embryos were aged until first instar [24 hour After Egg Laying (AEL)] and heat shocked for 30 minutes at 37°C. Larvae were aged at room temperature until late 3rd instar (about 120 AEL) and brains were dissected.
To generate larval brains without optic lobes, so-Gal4 (Chang et al., 2003) was used to drive the expression of a UAS-dominant negative-EGFR (Buff et al., 1998).
Immunohistochemistry and Imaging
Larval brains were dissected in PBS and fixed in PBT (PBS with 0.1% Triton X-100) containing 4% formaldehyde for 30 minutes at room temperature. Standard staining protocols were used (Ashburner, 1989). Primary antibodies were used at the following concentrations: mouse anti-Neurotactin (Hortsch et al., 1990; 1:10; Hybridoma Bank); mouse anti-Syntaxin (Fujita et al., 1982; 1:10; Hybridoma Bank); rat anti-DEcad (Dumstrei et al., 2003b; 1:500); mouse anti-acetylated tubulin (1:1000; Sigma T6793); mouse anti-β-galactosidase (1:50; Sigma); rat anti-DN-cadherin (Iwai et al., 1997; 1:20; Hybridoma Bank). Secondary antibodies were purchased from Jackson Laboratory and used at the manufacturer’s recommended concentrations. Stained brains were mounted in Vectashield (Vector Laboratory; H-1000).
Confocal images were taken on a Biorad MRC1024ES microscope using Laser sharp version 3.2 software. Complete series of optical sections were taken at 2µm intervals. Images were analyzed using the ImageJ software. Generation of three-dimensional models was done with the Amira 3.0 software (Mercury Computer System, Inc.).
Immunoprecipitation of proteins from larval brain
100 larval brains were dissected in ice-cold PBS and homogenized in 0.2 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 1 mM DTT, 1 mM PMSF). Proteins were immunoprecipitated according to the methods described in Dumstrei et al. (2002), using anti-β-cat antibody (Developmental Studies Hybridoma Bank) at 1:50 dilution. Each immunoprecipitation experiment was done in duplicates. Co-immunoprecipitated proteins were separated on 8% SDS-PAGE and transferred to Western blots. Blots were probed with anti-DE-cad (Dumstrei et al., 2003b) at 1:1000 dilution; anti-β-cat at 1:500 dilution and anti-α-catenin (Oda et al., 1993) at 1:500 dilution.
Electron Microscopy
Larval brains were dissected in cold PBS and fixed in 4% paraformaldehyde, 2.5% glutaraldehyde in 0.1M phosphate buffer, pH 7.3, for 24 hours at 4°C. The brains were post-fixed for 60 minutes with 1% osmium tetroxyde in 0.1M phosphate buffer, pH7.3, at 4°C. Specimens were washed several times with distilled water and dehydrated in graded acetone series at 4°C. Specimens were incubated in 1:3, 2:2, 3:1 Epon:acetone ratio for 2 hours, 3 hours, and overnight, respectively. Brains were transferred to unpolymerized Epon and incubated overnight. They were then transferred to molds, oriented and placed at 60°C for 16 hours to permit polymerization of Epon. Blocks were sectioned at 0.06µm and mounted on slot grids and treated with uranyl acetate and lead citrate.
Results
DE-cad is dynamically expressed at early stages of SAT axon elongation
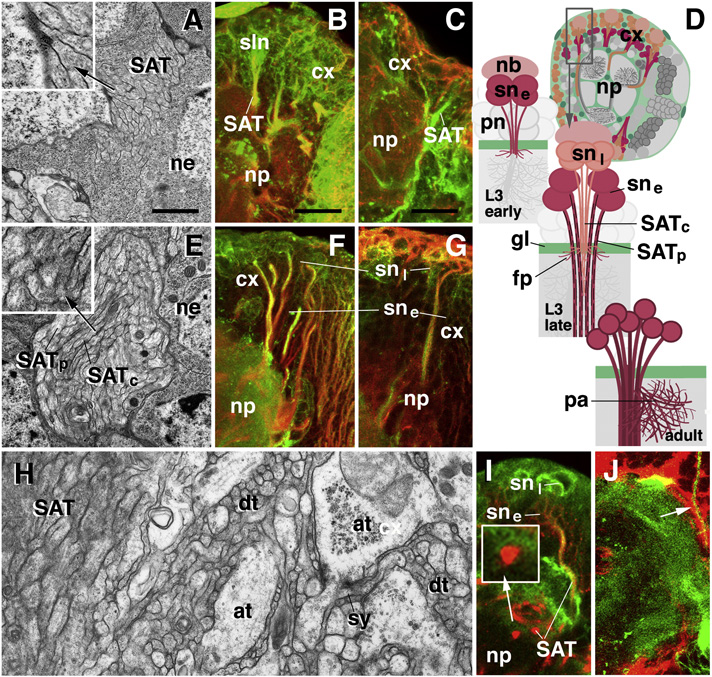
Secondary lineages of the larval brain are formed during the late second to late third instar. SATs grow into the neuropile compartments that had been formed by primary neurons during late embryonic and early larval stages. Each secondary lineage is defined by its position within the brain cortex and the invariant trajectory followed by its axons; based on these characteristics, lineages were mapped (Pereanu and Hartenstein, 2006). Secondary lineages share a number of generic properties. In most, if not all, lineages the sequence in which secondary axons form is reflected in their position within the SAT, and is also correlated with the expression of DE-cad and the organization of the cytoskeleton. As axons first grow out, they are thin, actin-rich, and express DE-cad (“early secondary axons”; Fig.1A–D). When reaching the glial layer that surrounds the neuropile surface (Fig.1J), these axons stall and grow out a tuft of filopodia. Subsequently, axons further elongate as a coherent bundle into the neuropile, often after following the neuropile surface for various distances. These older axons (“mature secondary axons”) become thicker and assemble more microtubules (Fig.1D–G). At the same time, the expression of DE-cad ceases (Fig.1F, G, I). Within the neuropile, SATs form bundles that consist exclusively of mature secondary axons that do not express DE-cad (Fig.1H, I). The axons forming later grow through the center of the pre-existing SAT tract (Fig.1D, middle). In other words, when looking at the cross section of a SAT (Fig.1E), older axons (thick, many MTs, DE-cad-negative) are always at the periphery of the tract, younger ones (thin, few MTs, DE-cad-positive) in the center. This peculiar manner of axon growth may enable older axons better to serve as “guides” for their younger siblings.
Fig. 1.
Development of secondary lineages in the Drosophila larval brain. A–C: Early third instar (72h after hatching). E–J: Late third instar (120h after hatching). D: Schematic diagram illustrating secondary lineage at early third larval stage (top), late third larval stage (middle), and adult (bottom). Panels on the left (A, E, H) are EM sections depicting secondary axon tracts in cross section. Right panels (B, C, F, G, I, J) are confocal sections showing DE-cad expression (green) in secondary lineages. Double label (red) is phalloidin for microfilaments (B, F), anti-Acetylated tubulin for microtubules (C, G), neurotactin for secondary lineages (I) and Nrv2-GFP for glial cells (J). At the early third instar, secondary axon tracts form bundles of 20–50 thin fibers traversing the cortex and reaching the cortex-neuropile boundary (A, D). These early secondary axons have the characteristics of filopodia, measuring in the order of 100nm in diameter, being rich in microfilaments, and lacking microtubules, (inset in A; B, C). All early fibers express DE-cad (B, C). At late third instar secondary axon tracts have grown in length, diameter and number of fibers (100–200). Fibers produced at earlier stage now measure 200–300nm and contain microtubules; these mature secondary axons are located at periphery of SAT (black arrow in E and inset E). Late born, immature fibers are in the center of the SAT (white arrow in E and inset of E); only these fibers express DE-cad. SATs of mature fibers have entered the neuropile where they stay together and are easily distinguishable from the adjacent primary neuropile (H, I). The mature SATs within the neuropile (labeled by anti-Neurotactin in I) have lower levels of DE-cad expression (inset in I). SATs are invested by glia in the cortex and at the cortex-neuropile boundary (J).
Abbreviations: at terminal axons of primary neuropile; cx cortex; dt dendritic terminals of primary neuropile; fp filopodia; gl glia; nb neuroblast; ne neuronal cell body; np neuropile; pa proximal arborization; pn primary neuron; SATc central (late born) fibers of SAT; SATp peripheral (early born) fibers of SAT; sne early born secondary neuron; snl late born secondary neuron; sy synapse
Scale bars: 1µm (A, E, H); 10µm (B, F, I); 5µm (C, G, J
Neuronally expressed DE-cad is diffusely expressed in the membrane and forms a complex with β-catenin and α-catenin
In the Drosophila embryo, DE-cad is expressed in epithelial cells where it interacts with β-cat and α-catenin (α-cat) (Tepass et al., 2000). The cadherin-catenin complex is strongly enriched in the zonula adherens, a narrow belt of adherens junctions encircling the apical pole of epithelial cells. By contrast, in neuroblasts and neurons of the larval brain, DE-cad is localized diffusely in the cell membrane. Electron microscopic analysis of larval brain cortex indicates that there are few, if any, adherens junction-type membrane specializations in between neuronal somata, or axons (Fig.1A, E, H; data not shown). However, cell membranes of SAT fibers are closely packed; the inter-cellular cleft in between fibers is less than 8nm (compared to the 20–25 nm cleft in between Drosophila epithelial cells).
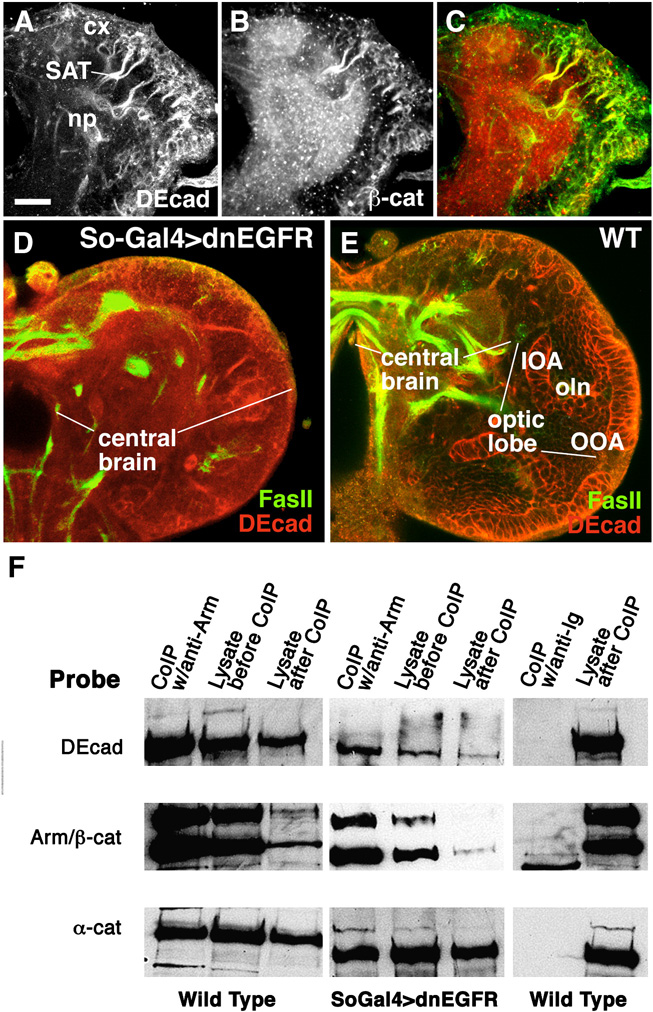
The DE-cad expression pattern in the secondary lineages of the larval brain coincides with the expression of its presumed binding partner, β-cat (Fig. 2A–C) and α-cat (data not shown). In addition to secondary neuroblasts and neurons, β-cat is expressed strongly in the neuropile; this most likely reflects the fact that β-cat also forms a complex with DN-cad, which is expressed on primary neurons forming in the neuropile. Given that few or no adherens junctions exist among neuronal somata and axons, we wondered whether DE-cad and the catenins form a regular protein complex as described for epithelia. To this end, co-immunoprecipitation (coIP) experiments were performed on protein extracts from late larval brains. In wild type, the epithelial optic anlagen constitute a major part of the brain (Fig.2E), which would affect the interpretation of our results. We therefore generated brains lacking optic anlagen by expressing a dominant negative construct of EGFR, in the embryonic optic lobe primordium. In both transgenic lines, larvae and adult flies were viable, but completely lacked optic lobe and eye (Fig.2D). CoIP of extracts of these brains confirmed that DE-cad, β-cat and α-cat form a protein complex, just as in epithelial cells (Fig.2F).
Fig. 2.
DE-cad expressed in the larval brain is associated with β-cat and α-cat. A–C: Confocal sections of early third instar larval brain labeled with anti-DE-cad and anti-β-cat antibody. DE-cad appears strongly in neuroblasts and early neurons with their axons (SAT) as well as the optic lobe anlagen. The same structures express β-Cat; in addition, the neuropile (np) is strongly positive. This expression most likely reflects the presence of catenin in terminal branches and synapses formed by primary neurons, which are negative for DE-cad but express DN-cad (not shown). D, E: Confocal section of late third instar brain in which outer and inner optic anlagen (OOA, IOA) of optic lobe were ablated by embryonic expression of so-Gal4/UAS-dn-EGFR; control section in E. F: Western blots containing Co-IP products. Left: Extract of wild-type larval brain co-immunoprecipitated using anti-β-cat. Blot is probed with anti-DE-cad, anti-β-cat and anti-α-cat, demonstrating that in the wild-type larval brain, DE-cad is associated with β-cat and α-cat. The same result is obtained when extract from optic lobe-less brains are co-immunoprecipated with the same probes, indicating that brain neurons and their processes also contain a cadherin-catenin complex. Blot shown in the right panel documents a control in which extract of wild-type larval brain is co-immunoprecipated with a mouse IgG.
Bar: 25µm
Loss of DE-cad and β-cat results in excessive branching and abnormal trajectories of SATs at the cortex-neuropile boundary
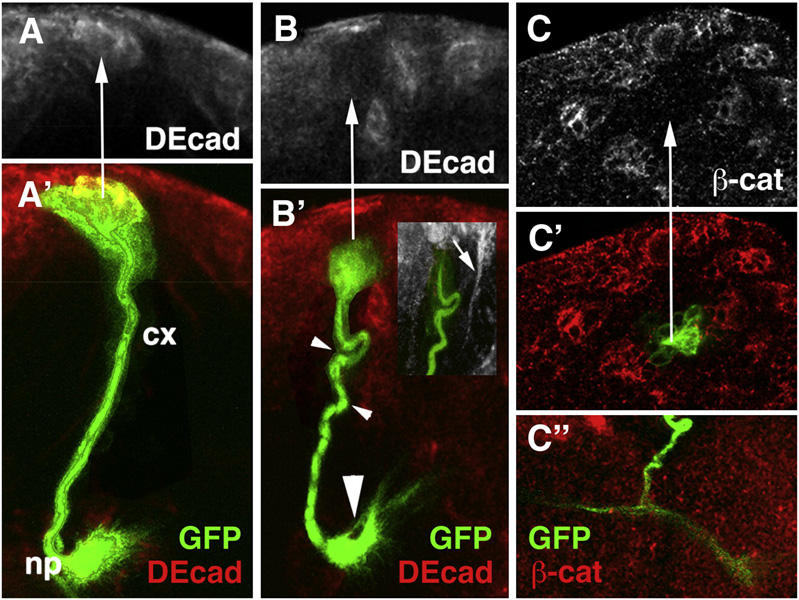
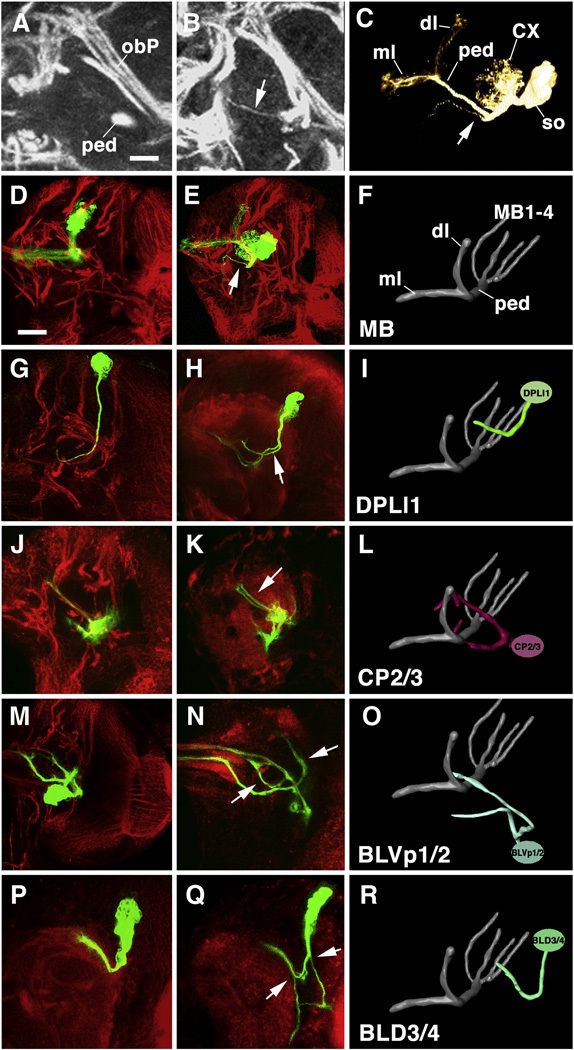
Utilizing the MARCM technique (Lee and Luo, 1999), ectopic GFP expression was induced in individual neuroblasts in the larval brain. Heat-pulses inducing somatic recombination were applied during the early larval stage, so that only secondary lineages (i.e., neurons born in the larval period), consisting of neuroblasts, GMCs, and neurons were visualized by the presence of the GFP protein. The morphological characteristics of a GFP-labeled wild-type control clone are shown in Fig.3A. The neuroblast is located at the surface of the cortex. Clusters of 3–4 GMCs, distinguishable from neurons by their larger size and reduced Elav expression (not shown), cling to the lateral surface of the neuroblast. Secondary neurons typically form a wedge-shaped or cylindrical cluster that reaches from the surface to close to the cortex-neuropile boundary (Fig.3A’). The axons formed by all of the neurons of one lineage remain as a tight bundle, the SAT, that projects towards the neuropile, penetrates the glial layer at the cortex-neuropile boundary, and travels for various distances within the neuropile. Approximately 30% of the lineages split into two branches with different trajectories at the cortex-neuropile interface; about 10% split into three branches.
Fig. 3.
Elimination of DE-cad by the MARCM technique can be confirmed by reduced levels of expression of DE-cad and β-cat. A, A’: Confocal section of wild-type MARCM clone (green) corresponding to BLD3/4 lineage. Note strong DE-cad expression in cell bodies (arrow) in cortex. B, B’, C, C’: DE-cad loss-of-function clones. Note loss of DE-cad expression (arrow in B) and β-cat expression (arrow in C); note also abnormal SAT trajectories (small arrowhead in B’) and excessive branching at cortex-neuropile boundary (large arrowhead in B’ and C”).
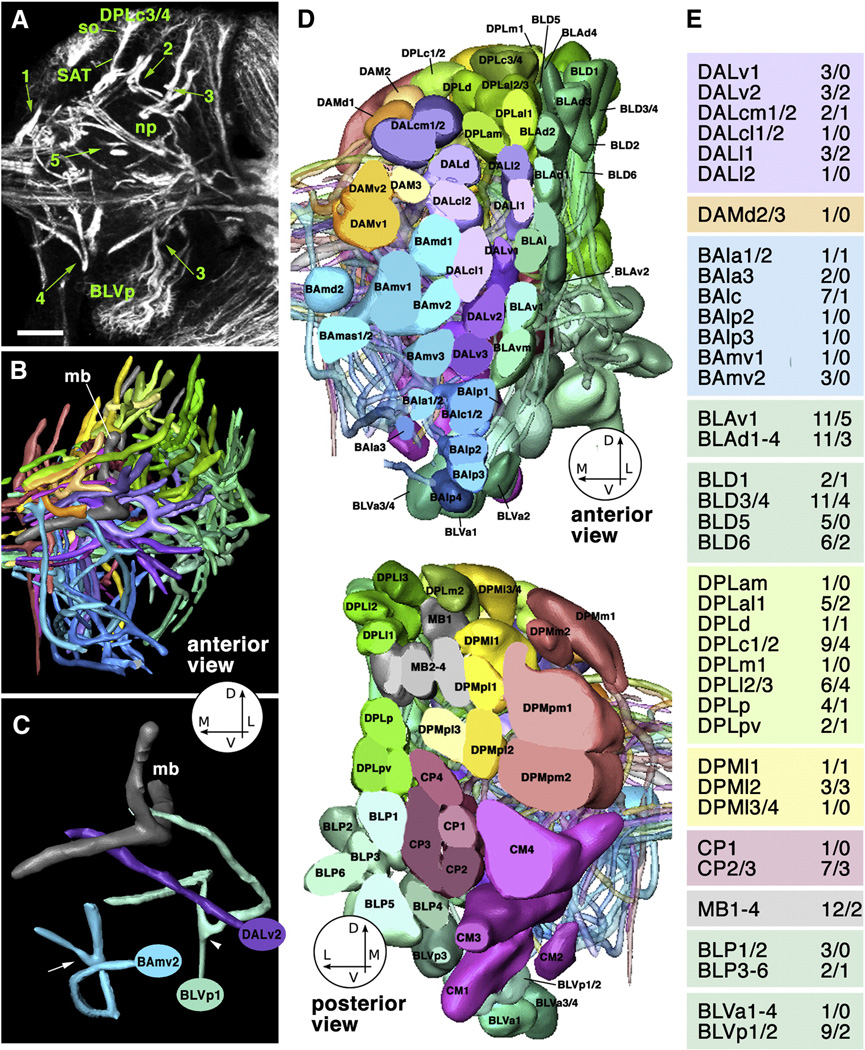
DE-cad mutant clones showed absence or significant reduction of DE-cad expression in cell bodies and axons (Fig.3B, B’). The structural abnormalities of these clones included changes in cell body position and axon defasciculation, but most prominently, an increased number of branches at the cortex-neuropile boundary, as well as abnormal SAT trajectories in the neuropile. Figure 4 summarizes the characteristics of brain lineages that can be scored when identifying mutant phenotypes. All lineages are characterized by a highly stereotyped SAT pattern. This pattern includes (1) the point of entry into the neuropile; (2) direction of SAT at cortex-neuropile boundary (SAT follows straight radial path into neuropile or turns and follows neuropile surface); (3) presence or absence of SAT branching at the cortex neuropile boundary; (4) fasciculation of SAT with other tracts in the neuropile; (5) branching of SAT in the neuropile. Figure 4A and C give examples SATs branching at cortex-neuropile boundary (e.g., BLVp1/2) or within neuropile (e.g., BAmv2). Using these criteria, we can reliably recognize almost all lineages individually (Fig.4D). In several cases (e.g., DALcm1/2; CP2/3), lineages form pairs in which clusters of cell bodies lie next to each other and produce separate SATs, but then merge into one single tract with a common trajectory and target. Five lineages (DPMm1, DPMpm1 and DPM2, CM3, CM4) stand out by their much larger size (Fig.4D, bottom panel). These so-called “mega-lineages” follow a different proliferatory pattern (Bello et al., 2008), in that the neuroblast first produces several symmetrically dividing “amplifying progenitors”, whose daugher cells then behave like regular neuroblasts. The mega-lineages have 4–5 times more neurons than regular lineages, and produce complex arrays of SATs. Since the pattern of the SATs of mega-lineages is not yet fully worked out, clones in these lineages were not considered in the present study.
Fig. 4.
Topology and structural characteristics of wild-type lineages of the late larval brain. A: Z-projection 10 successive 1µm confocal cross sections at level of central neuropile, Secondary lineages, their axon tracts (SATs) and neuropile fascicles formed by convergence of SATs are labeled with anti-Neurotactin antibody (white). Clusters of somata (so) belonging to lineages are located in the cortex; axon tracts project centripetally into the neuropile (np). Green arrows and numbers indicate points of interest concerning SAT trajectory: (1) SAT proceeds straight from cortex into neuropile (DPMm1 lineage); (2) SAT makes sharp turn when reaching the cortex-neuropile boundary (DPLm1 lineage); (3) SAT bifurcates into two branches at cortex-neuropile boundary (top: DPLl1/2; bottom: BLVp1/2; (4) distal part of SAT bifurcates in neuropile (BAmv2); (5) SATs run close to each other without merging (distal portion of DPLam and peduncle). B: 3D digital model of all SATs of one brain hemisphere, anterior view. Coloring of SATs follows a scheme used consistently in this and the following figures; all lineages belonging to one topological family are rendered in the same color. Circle with arrows in this and other panels indicate directions (D dorsal; L lateral; M medial; V ventral). Dorsal lobe and medial lobe of mushroom body (mb) are shaded gray. C: Digital model of three lineages illustrating typical branching behavior of SATs [DALv2: straight unbranched entry into neuropile; BLVp1: bifurcation at point of entry into neuropile (arrowhead); BAmv2: bifurcation in distal leg of SAT (arrow)]. D: 3D digital models of all clusters of neuronal somata representing all lineages of one brain hemisphere. Top: anterior view; bottom: posterior view. For both models, the polar region of the cortex was “sliced off” to allow for clearer view of lineages. Lineages are annotated according to Pereanu and Hartenstein (2006). E: List of lineages for which DEcad mutant clones were generated and analyzed. First number indicates clones observed; second number refers to clones with detectable SAT abnormalities.
Bar: 25µm.
The frequency of clones with and without morphological phenotypes are listed in Fig.4E. Overall 43 out of 134 clones (32%) showed abnormalities. If we compared the frequency with which individual lineages were affected (Fisher exact analysis) to the overall score, none of the scores were significantly lower or higher than the overall 32%, due to the relatively small n-value. If we grouped lineages by their topology (e.g., all BA, DPL, DPM etc added together), some significant differences materialized (Tab.1).The grouping of lineages in families is based on a common primary target. Thus, for example, all lineages whose SAT enter the neuropile in or directly adjacent to the BA compartment (the larval antennal lobe) are designated as “BA lineages” (Pereanu and Hartenstein, 2006).
Table 1.
Statistical analysis of the frequency of abnormalities in DE-cad mutant clones.
| A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All Clones | DAL | DAM | BA | BLA | BLD | DPL | DPM | CP | MB | BLV | |
| Total clones | 134 | 13 | 1 | 16 | 22 | 13 | 29 | 5 | 8 | 12 | 10 |
| Abnormal clones | 43 | 5 | 0 | 2 | 8 | 3 | 13 | 4 | 3 | 2 | 2 |
| Percent of Total | 32% | 38% | 0% | 13% | 36% | 23% | 45% | 80% | 38% | 17% | 20% |
| p-value | n/a | 0.210 | 0.681 | 0.066 | 0.175 | 0.209 | 0.072 | 0.041 | 0.277 | 0.154 | 0.221 |
| B | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DAL | ||||||||||
| DAM | 0.643 | |||||||||
| BA | 0.099 | 0.882 | ||||||||
| BLA | 0.279 | 0.652 | 0.081 | |||||||
| BLD | 0.236 | 0.786 | 0.289 | 0.219 | ||||||
| DPL | 0.247 | 0.567 | 0.023 | 0.189 | 0.117 | |||||
| DPM | 0.132 | 0.333 | 0.011 | 0.092 | 0.045 | 0.145 | ||||
| CP | 0.354 | 0.667 | 0.158 | 0.328 | 0.295 | 0.295 | 0.163 | |||
| MB | 0.176 | 0.846 | 0.387 | 0.161 | 0.355 | 0.071 | 0.027 | 0.238 | ||
| BLV | 0.236 | 0.818 | 0.361 | 0.223 | 0.382 | 0.121 | 0.045 | 0.294 | 0.406 | |
| DAL | DAM | BA | BLA | BLD | DPL | DPM | CP | MB | BLV |
Calculations were carried out using “Simple Interactive Statistical Analysis” available at http://www.quantitativeskills.com/sisa/statistics/t-thlp.htm. Because the data set contained values close to or equal to 0, Fisher exact analysis was used for all comparisons. Design effects were set to 1.0, and confidence intervals were set to 95%. An exact p-value less than or equal to 0.05 was considered significant.
(A) To find whether topologically defined lineage families (e.g., BA DPM) differ in their requirement for DE-cad function, the number of all clones belonging to a given family was divided by the number of all clones with abnormalities in that family; this provided the “family score”, which then was compared to the overall score (all clones added). Families that show a significant difference from the overall score are highlighted in red. In (B), family scores are compared to each. As in (A), all significant differences are highlighted in red.
Note: 5 BLP-family clones containing 1 abnormal clone are not shown in these tables.
Comparing the ratio of abnormal clones that belong to a given lineage family to the overall score, one family, DPM, stands out by having a particularly high number of clones with abnormal phenotypes. Comparing lineage families to each other showed that BA lineages (antennal projection neurons and other lineages whose SATs project into the ventro-anterior brain) showed abnormal phenotypes less often than DPL and DPM lineages (located in the dorsal brain). Similarly, mushroom body lineages showed significantly fewer clones with abnormalities than DPM lineages, and marginallly fewer than DPL. The data imply that the requirement for DE-cad varies: in terms of SAT branching and trajectory, some lineages are more resistant to the loss of DE-cad than others. This conclusion was further supported by the result of an experiment where we used the atonal-Gal4 driver (Hassan et al., 2000), expressed in BLD5, to generate and visualize more clones specifically in this lineage. Out of more than 15 clones overall, not a single one showed ectopic branches or fasciculation defects.
Figure 5 illustrates typical structural phenotypes encountered. A–F show an MB lineage where the SAT (forming part of the peduncle) gives off a side branch within the neuropile, near the beginning of the peduncle. More frequently than ectopic branches in the neuropile we encountered abnormalities starting at the cortex neuropile boundary. Shown in Fig.5G–I is a DPLl1 lineage whose normally single SAT splits into two at the point where it enters the neuropile; both branches then follow a similar trajectory. A similar phenotype can be seen for the clone affecting the CP2/3 lineage (Fig.5J–L). CP2/3 form a pair of lineages whose SAT normally splits into a dorsal branch, crossing over the peduncle, and a ventral branch, traveling first parallel to the peduncle and then converging onto the spur region (Fig.5L). In the mutant clone, the dorsal branch is divided into two components traveling close to each other, but separately, a behavior never encountered in a wild type CP2/3 clone. The examples shown in Fig.5M–O and P–R illustrate cases where supernumerary branches follow different trajectories. The BLVp1/2 clone, normally branched into two long SATs (Fig.5M, O) forms two extra branches that follow trajectories not seen in any wild-type lineages (arrows in Fig.5N). Similarly, the mutant BLD3/4 clone shown in Fig.5P–R generates two supernumerary branches at the cortex-neuropile boundary with novel trajectories (arrows): projecting ventro-laterally this branch reaches the optic lobe neuropile, where it forms a tuft of filopodia, before turning sharply medially to join a commissural tract. A second ectopic branch projects straight ventrally towards the ventral nerve cord.
Fig. 5.
Phenotypic abnormalities in SAT branching and trajectory of DE-cad mutant clones. Panels of the middle column show Z-projections of frontal confocal sections of DE-cad mutant MARCM clones, paired up with Z projections of their wild-type counterparts (left column), and 3D renderings of these wild-type lineages (right column). In confocal images, clones are labeled green (GFP); global marker for lineages (anti-Neurotactin) or neuropile (anti-DNcad) was used as counterstaining (red), In panels of this and the following figure, only the right brain hemisphere is shown; the brain midline coincides with the left panel margin. 3D models are shown from antero-dorsally; the mushroom body is included for orientation. Arrows point at ectopic branches of SATs. A–F: Clone in mushroom body lineage. Note ectopic branch extending from SAT in proximal peduncle (ped; B, C, E). In wild type (A, D), no side branches of peduncle are ever observed. Panel C shows volume rendering of the DE-cad mutant mushroom body clone; anterior to the left (CX calyx; dl dorsal lobe; ml medial lobe; ped peduncle; so cluster of somata). G–I: Clone in DPLl1 lineage; J–L: CP2/3 lineage; M–O: BLVp1/2 lineage; P–R: BLD3/4 lineage.
Bars: 5µm (A, B); 25µm (all other panels).
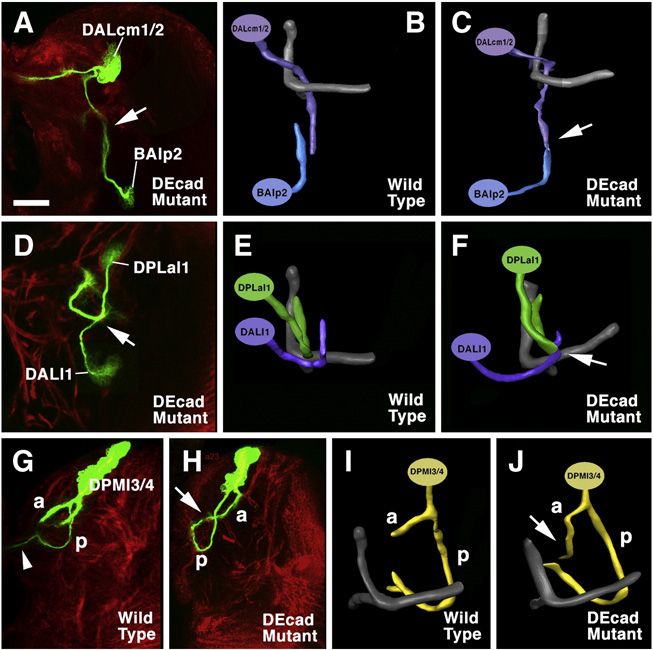
An additional aspect of SAT pathway abnormalities within the neuropile was revealed in brains that contained several mutant clones together. In such cases, it could be observed that SATs that in wild type remain separate throughout their pathway, fused in the mutant clones. Shown in Fig,6A–C is DALcm1/2 and BAlp2. One branch of DALcm1/2, in wild type, turns ventrally and forms a tract towards the ventral nerve cord. BAlp2 has an upward directed, unbranched SAT. Both come within less than 5µm from each other, but do not fasciculate (Fig.6B). In brains where both lineages lacked DE-cad, the SATs merged (Fig.6C, E). A second example is the pair DPLal1 and DALl1. In wild type, these form characteristic crescent shaped SATs that arch around each other without touching (Fig.6E). In the mutant setting, the SATs fuse at the point of closest proximity (Fig.6D, F). The phenotype shown by the mutant clone in DPMl3 (Fig.6G–I) shows similarities to the fusions above, although only a single lineage is involved here. Thus, the wild-type DPMl3 splits into two branches at the point where it enters the neuropile: an antteriorly directed branch (“a” in Fig.6G, I) and a posteriorly directed one (“p”) The latter curves ventrally and medially and splits into a commissural component (arrowhead in Fig.6G, I) and a short ipsilateral branch that ends close to the commissure. The posterior branch of the mutant DPMl3 (Fig.6H, J) initially follows the same pathway as its wild-type counterpart; however, it does not enter the commissure, but turns backward and terminates in contact with the tip of the anterior branch of the same lineage, thereby forming a closed circular SAT.
Fig. 6.
SAT convergence in DE-cad mutant clones. Panels on the left (A, D, H) are Z-projections of frontal confocal sections of GFP labeled mutant clones (green); anti-Neurotactin (global marker for secondary lineages) is used as counterstaining (red). Panels of the right column (C, F, J) are 3D digital models of the clones; panels in center (B, E, I) show models of corresponding wild-type lineages. A–C: Convergence of DALcm1/2 and BAlp2. In wild-type, SATs of these lineages grow towards each other from dorsally and ventrally, but pass without coming into contact (B); the SATs merge in the mutant double clone (arrows in A and C). D–F: the same phenomenon is shown for DPLal1 and DALl1. G–J: Clone in DPMl3/4 lineage. SAT bifurcates into anterior (a) and posterior (p) branch; in wild-type, p projects across the midline (arrowhead). In mutant, p converges upon the a branch (arrow in H and J).
Bar: 25µm
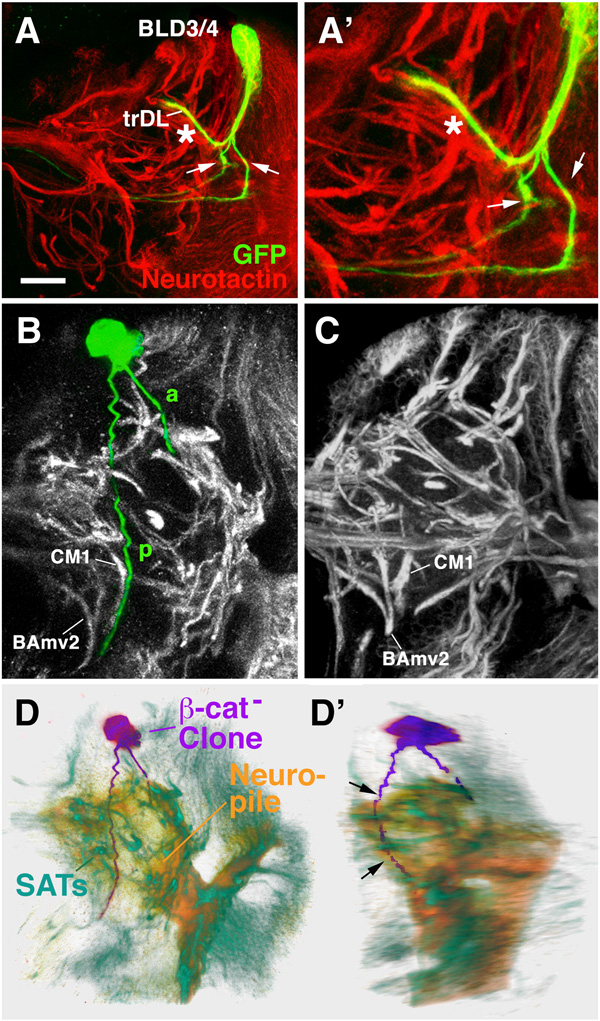
β-cat is the cytoplasmic binding partner of DE-cad and shows an overlapping expression in secondary lineages and their SATs (see above). Correspondingly, loss of function MARCM clones in β-cat have similar phenotypes as DE-cad clones (Fig.7). The abnormalities included mostly excessive branching at the cortex-neuropile boundary, as shown for the BLD3/4 lineage depicted in Fig.7A. Within 43 total clones, we observed several clones whose SAT did not enter the neuropile at all, but skirted along the neuropile surface (Fig.7B–D). Cell bodies of the clone shown here are located in the DPMl territory. The course taken by the SAT also have similarity to the DPMl3/4 pattern (see Fig.6G–J above). One branch grows postero-ventrally, the other anteriorly. However, both branches remain superficially at the neuropile surface and deviate from their normal pattern. The posterior branch does not enter the commissure but continues stright downward towards the ventral nerve cord. The anterior branch terminates at a superficial position near the dorsal lobe of the mushroom body.
Fig. 7.
Loss of β-cat leads to defects in axonal trajectory of secondary lineages. A–A’: Z projections of two β-cat mutant MARCM clone representing the BLD3/4 lineages. In wild-type, BLD3/4 lineages have one branch (see Fig.5R) that makes a sharp turn into the trDL tract. Aside from this branch which is indicated by asterisk in A – A’, the two mutant clone forms two additional branches following extraneous pathways (arrows). These extra branches appear to express no or low levels of neurotactin. B: Z projections of β–cat mutant MARCM clone representing DPMl lineage. Based on SAT trajectory, showing a split into a forward (a) and back branch (p), this lineage resembles most closely the wild-type DPMl3/4 (see Fig.6G, I). However, the posterior branch does not cross the midline as in wild type, but rather continues ventrally in close proximity to the SAT of CM1. C shows Z projection of wild-type brain at comparable level to that in B. Note characteristic SATs of BAmv2 and CM1 in both B and C; in wild type, no SAT appears at the position of the p branch of the mutant clone. D, E: Volume renderings of the β-cat loss-of-function clone shown in panel B in frontal view (C) and latero-frontal view (D). The clone (purple) is shown against the background of a volume rendered brain neuropile (orange) and anti-Neurotactin-labeled SATs (green). Note that the SAT (both branches) of the labeled clone does not follow any of the normal (neurotactin-positive) SATs into the neuropile, but skirts the posterior neuropile surface on its abnormal course ventrally (arrows in D’).
Bar: 25µm
An interesting phenotype shown by several of the β-cat mutant clones concerned a downregulation of the global neuronal marker, neurotactin. In wild type, neurotactin is expressed on all secondary neurons and their axons from the time they grow out. Mutant clones often showed branches that had lost neurotactin expression (Fig.4A’, A”). In these clones, cell bodies and the proximal segment of the SAT (within the cortex) were positive for neurotactin, but one or more of the supernumerary branches penetrating into the neuropile were neurotactin-negative. This finding points at regulatory relationships between the different adhesion complexes formed by DE-cad and neurotactin.
Discussion
Cadherins are structural membrane molecules expressed throughout the developmental history of most, if not all, animal cell types. Most cells express multiple cadherins in a dynamic pattern, such that one cadherin is turned on at an early stage and is later replaced by another cadherin. Given their widespread expression it is no wonder that cadherins participate in many different functions, both developmentally and in the mature organism. In the larval Drosophila brain, the reduction of DE-cad by expression of a dominant-negative construct results in abnormalities of neuroblast proliferation, glial morphology, layering of neurons, and axon patterning (Dumstrei et al., 2003a). Abnormalities in a wide range of morphogenetic processes have also been observed in vertebrate. In zebrafish, hypomorphic mutation of E-cadherin showed flattened anterior neural tissue, scattered trigeminal ganglia, and caused abberant axon bundles from the trigeminal ganglia (Shimizu et al., 2005). In mouse, loss of E-cadherin (Cdh1) during neurogenesis results in loss of polarity and adhesion among radial glial cells, disrupting the integrity of the ventricular zone (Rasin et al., 2007). How can one envision, in more general terms, the cellular mechanism for which cadherin function is essential? Studies carried out so far point at a number of different actions of cadherins.
(1) Differential expression of cadherins is involved in specifying brain compartments and neural circuits: it has been known for a long time that boundaries between tissues are set up by cell sorting (Townes and Holtfreter, 1955). If cells within one population start expressing two different adhesion molecules (or different levels of the same molecule) they will sort, such that cells with the same adhesion molecule will group together. It is widely speculated that the fact that cadherins are expressed in highly diverse and regionally specific patterns in the developing vertebrate nervous system points at a role of these molecules in “neural sorting”, in the sense that neurons or axons sort out into specific brain compartments or tracts depending on the type of cadherin they express (Redies, 2000; Tepass et al., 2000; Yagi and Takeichi, 2000). Experimental evidence for the role of cadherins in sorting neuronal connectivity by matching discrete populations of neurons with their targets comes from studies of the visual system. In the chicken optic tectum, N-cadherin is concentrated at laminae b–e. If N-cadherin is blocked by antibodies, retinal axons fail to stop at these target laminae (Sanes and Yamagata, 1999). In a similar manner, DN-cad may regulate target specifity in visual and olfactory sensory afferents (Hummel and Zipursky, 2004; Lee et al., 2001; Zhu and Luo, 2004).
(2) Cadherin-mediated adhesion modulates signaling pathways, or plays a permissive role during cell signaling interactions: Numerous examples of cases where the cadherin-mediated contact between cells activates specific signaling cascades are known (e.g., (Williams et al., 2001; Yap and Kovacs, 2003). In addition, a permissive effect of cadherins on signaling can be envisaged whereby cell contacts at a certain time or space need to be established/stabilized in order for specific signal-receptor interactions to occur.
We speculate that it is this second role of cadherins that most likely explains the axonal abnormalities that we see in DE-cad loss of function clones. As the first secondary axons start to extend as a cohesive bundle (SAT), they have to grow only for a short distance before reaching the glial layer surrounding the neuropile. After traversing this layer the SAT continues to extend, either entering the neuropile immediately, or by growing for various distances, and in different directions, along the glial layer (Dumstrei et al., 2003b; Pereanu and Hartenstein, 2006). Since lineages differ with respect to the trajectory of their SAT, one has to assume that specific signals expressed locally within the lineage and/or their environment guide the choice of the SAT which direction to follow. The fact that SATs of DE-cad mutant lineages make the wrong choice in a significant fraction of cases indicates that DE-cad-mediated adhesion, possibly between axons and neuropile glia, or between axons themselves, is required at the “choice point” to receive the proper “instructions”. We prefer this permissive model of DE-cad function to an instructive model because DE-cad expression on SATs and neuropile glia appears to be uniform for all lineages, despite the fact that lineages differ widely in their pathway choices.
It is important to point out that the requirement of different lineages for DE-cad is apparently not uniform. Thus, for some lineages (example: most BA lineages; BLD5), abnormalities were never observed in mutant clones; in other cases, the opposite was true, and mutant clones often caused abnormal pathway choices. This finding suggests that the specific cues determining the pathway choice of a given SAT are more “robust” for some lineages, promoting the right choice even in the absence of permissive factors such as DE-cad. The finding also underscores the importance of assaying many lineages, rather than one, when evaluating the function of a gene in neural development. Negative findings in a genetic analysis that focuses on one (or a small group of lineages, such as the mushroom body, which has been so far used almost exclusively as a “testing ground” for genetic studies on brain development) do not imply that the gene in question plays no role in other neurons. Looking at different lineages in the Drosophila larval brain has only recently become possible, since the morphology, different for each one of the 100 or so lineages, has been described (Pereanu and Hartenstein, 2006).
The DE-cad loss of function phenotype that we observe in clones is relatively mild, compared to the findings in a previous study where we utilized a dominant negative construct to inhibit DE-cad in larval brain development (Dumstrei et al., 2003a). First, the mutant clones reveal no gross defects in size or neuronal position. We had seen such defects in experiments where a dominant negative DE-cad construct was driven, either in the entire brain (neurons plus glia), or in glia alone. In the latter case we could verify directly that the glial scaffold (“trophospongium”) around neuroblasts and neurons, formed by the widespread processes of cortex glia, is absent. Taken together, the findings suggested that DE-cad mediates interactions among cortex glia which are required for the formation of the trophospongium. Once in place, the trophospongium is able to support proper neuroblast division and neuronal layering, even if DE-cad is removed from the neurons by somatic recombination. Another possibility to keep in mind is perdurance, where the DE-cad inherited from the neuroblast prior to clone induction, is sufficient to function in early processes like neuroblast division and layering of neurons.
The DE-cad LOF phenotype in mutant clones is also quite mild with respect to axon fasciculation and pathfinding. Pathway abnormalities were observed in a substantial fraction of, but not all, cases; only rarely did individual axons within the SAT of a clone defasciculate before entering the neuropile. Here it is important to keep in mind that aside from DE-cad, many additional adhesion molecules are expressed in developing neurons. For example DN-cad and neurotactin: DN-cad is expressed at a low level in differentiating neurons, and starts to be upregulated in the late larva. Neurotactin is expressed in all secondary neurons from the time onward when they are born. It is plausible that all three proteins, in addition to other adhesion molecules (Fung et al., 2008), would share in the function of mediating contact between axons/axon-glia at the point where the SAT enters the neuropile and “searches for” specific cues guiding its further progress into the neuropile.
Our findings indicate that β-cat, similar to its role in epithelial morphogenesis, interacts with DE-cad in the developing nervous system. Loss of β-cat in individual clones results in phenotypes similar to those, if not often stronger, than those observed in DE-cad mutant clones. Many studies in the vertebrate field support a role of β-catenin and other proteins that bind to the cadherin-catenin complex in neural development, and particular in synaptic target selection, synaptogenesis and synaptic function (Elul et al., 2003; Murase et al., 2002; Togashi et al., 2002; Yu and Malenka, 2004). It has been speculated that the role of β-catenin in axons is similar to that in epithelial cells, namely to mediate the dynamic interaction between membrane adhesion complexes and the actin cytoskeleton (Thoumine et al., 2006). It will be important to learn more about the molecular basis of this link in growing axons. Axons and their growth cones navigate in a three-dimensional space; their interaction with the microenvironment is extremely dynamic, judging from the live recordings of filopodia that extend from progressing growth cones. This is different from epithelial cells which form two dimensional sheets, and generally move in concert. One might expect differences in the mechanism that integrate cell-contact (adhesion complex) and cell movement (cytoskeleton). Genetic screens and in vivo studies in the Drosophila brain provide a useful tool to make progress in understanding this mechanism.
Acknowledgments
We are grateful to Dr. RW Carthew for providing us with fly stocks. This work was supported by NIH Grant RO1 NS29357-15 to V.H. and the Ruth L. Kirschstein National Research Service Award GM007185 to S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M. Drosophila, A Laboratory Manual. New York: Cold Sprint Harbor Laboratory Press; 1989. [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;19:3–5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buff E, Carmena A, Gisselbrecht S, Jimenez F, Michelson AM. Signalling by the Drosophila epidermal growth factor receptor is required for the specification and diversification of embryonic muscle progenitors. Development. 1998;125:2075–2086. doi: 10.1242/dev.125.11.2075. [DOI] [PubMed] [Google Scholar]
- Chang T, Younossi-Hartenstein A, Hartenstein V. Development of neural lineages derived from the sine oculis positive eye field of Drosophila. Arthropod Struct Dev. 2003;32:303–317. doi: 10.1016/j.asd.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Hartenstein V. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J Neurosci. 2003a;23:3325–3335. doi: 10.1523/JNEUROSCI.23-08-03325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Nassif C, Hartenstein V. Early development of the Drosophila brain: V. Pattern of postembryonic neuronal lineages expressing DE-cadherin. J Comp Neurol. 2003b;455:451–462. doi: 10.1002/cne.10484. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Shy D, Tepass U, Hartenstein V. Interaction between EGFR signaling and DE-cadherin during nervous system morphogenesis. Development. 2002;129:3983–3994. doi: 10.1242/dev.129.17.3983. [DOI] [PubMed] [Google Scholar]
- Elul TM, Kimes NE, Kohwi M, Reichardt LF. N- and C-terminal domains of beta-catenin, respectively, are required to initiate and shape axon arbors of retinal ganglion cells in vivo. J Neurosci. 2003;23:6567–6575. doi: 10.1523/JNEUROSCI.23-16-06567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S, Wang F, Chase M, Godt D, Hartenstein V. The expression domains the cadherins in the Drosophila larval brain. J Comp Neur. 2008;506:469–488. doi: 10.1002/cne.21539. [DOI] [PubMed] [Google Scholar]
- Fujita SC, Zipursky SL, Benzer S, Ferrus A, Shotwell SL. Monoclonal antibodies against the Drosophila nervous system. Proc Natl Acad Sci U S A. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan BA, Bermingham NA, He Y, Sun Y, Jan YN, Zoghbi HY, Bellen HJ. atonal regulates neurite arborization but does not act as a proneural gene in the Drosophila brain. Neuron. 2000;25:549–561. doi: 10.1016/s0896-6273(00)81059-4. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Carthew RW. Surface mechanics mediate pattern formation in the developing retina. Nature. 2004;431:647–652. doi: 10.1038/nature02952. [DOI] [PubMed] [Google Scholar]
- Hill E, Broadbent ID, Chothia C, Pettitt J. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol. 2001;305:1011–1024. doi: 10.1006/jmbi.2000.4361. [DOI] [PubMed] [Google Scholar]
- Hirano S, Suzuki ST, Redies C. The cadherin superfamily in neural development: diversity, function and interaction with other molecules. Front Biosci. 2003;8:d306–d355. doi: 10.2741/972. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traquina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Hummel T, Zipursky SL. Afferent induction of olfactory glomeruli requires N-cadherin. Neuron. 2004;42:77–88. doi: 10.1016/s0896-6273(04)00158-8. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Hirota Y, Ozaki K, Okano H, Takeichi M, Uemura T. DN-cadherin is required for spatial arrangement of nerve terminals and ultrastructural organization of synapses. Mol Cell Neurosci. 2002;19:375–388. doi: 10.1006/mcne.2001.1081. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Murase S, Mosser E, Schuman EM. Depolarization drives beta-Catenin into neuronal spines promoting changes in synaptic structure and function. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. J Comp Neurol. 2003;455:417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- Oda H, Uemura T, Shiomi K, Nagafuchi A, Tsukita S, Takeichi M. Identification of a Drosophila homologue of alpha-catenin and its association with the armadillo protein. J Cell Biol. 1993;121:1133–1140. doi: 10.1083/jcb.121.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Jennett A, Younossi-Hartenstein A, Hartenstein V. A development-based compartmentalization of the Drosophila central brain. 2009 doi: 10.1002/cne.22376. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat Neurosci. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Redies C. Cadherins in the central nervous system. Prog Neurobiol. 2000;61:611–648. doi: 10.1016/s0301-0082(99)00070-2. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Yamagata M. Formation of lamina-specific synaptic connections. Curr Opin Neurobiol. 1999;9:79–87. doi: 10.1016/s0959-4388(99)80010-5. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Yabe T, Muraoka O, Yonemura S, Aramaki S, Hatta K, Bae YK, Nojima H, Hibi M. E-cadherin is required for gastrulation cell movements in zebrafish. Mech Dev. 2005;122:747–763. doi: 10.1016/j.mod.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Tepass U, Truong K, Godt D, Ikura M, Peifer M. Cadherins in embryonic and neural morphogenesis. Nat Rev Mol Cell Biol. 2000;1:91–100. doi: 10.1038/35040042. [DOI] [PubMed] [Google Scholar]
- Thoumine O, Lambert M, Mege RM, Choquet D. Regulation of N-cadherin dynamics at neuronal contacts by ligand binding and cytoskeletal coupling. Mol Biol Cell. 2006;17:862–875. doi: 10.1091/mbc.E05-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Townes PL, Holtfreter J. Directed movement and selective adhesion of embryonic amphibian cells. J Exp Zool. 1955;128:53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Wang F, Dumstrei K, Haag T, Hartenstein V. The role of DE-cadherin during cellularization, germ layer formation and early neurogenesis in the Drosophila embryo. Dev Biol. 2004;270:350–363. doi: 10.1016/j.ydbio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Williams G, Howell FV, Skaper SD, Walsh FS, Doherty P. Identification of an N-cadherin motif that can interact with the fibroblast growth factor receptor and is required for axonal growth. J Biol Chem. 2001;276:43879–43886. doi: 10.1074/jbc.M105876200. [DOI] [PubMed] [Google Scholar]
- Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- Yap AS, Kovacs EM. Direct cadherin-activated cell signaling: a view from the plasma membrane. J Cell Biol. 2003;160:11–16. doi: 10.1083/jcb.200208156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Yu X, Malenka RC. Multiple functions for the cadherin/catenin complex during neuronal development. Neuropharmacology. 2004;47:779–786. doi: 10.1016/j.neuropharm.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Zhu H, Luo L. Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron. 2004;42:63–75. doi: 10.1016/s0896-6273(04)00142-4. [DOI] [PubMed] [Google Scholar]